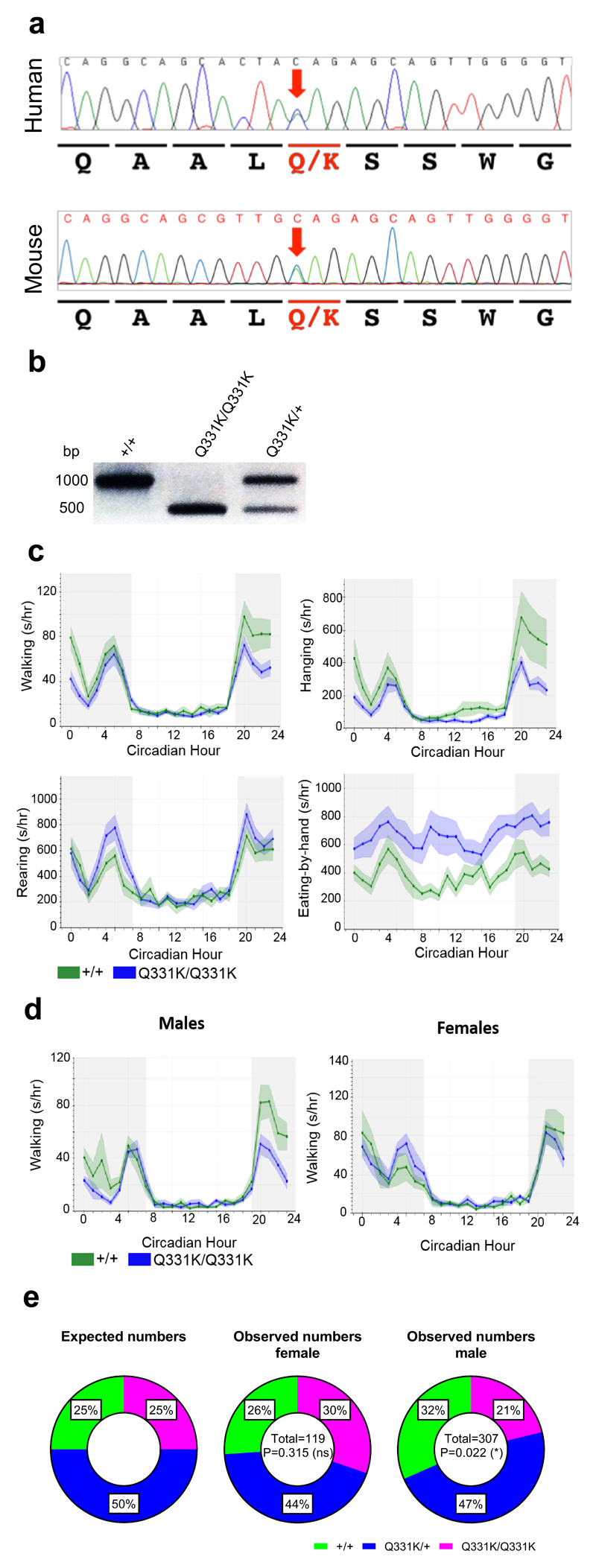
Figure 1. CRISPR mutagenesis, ACBM characterisation and breeding ratios of TDP-43Q331K mice.
(a) Chromatograms from the patient originally identified with the Q331K mutation and CRISPR/CAS9 knock-in founder mouse #52. Bases are given above the chromatograms and amino acids coded are given below. The mutation is highlighted with the red arrow.
(b) SapI restriction enzyme digestion of 1000 bp PCR products across the mutation site from representative genotyping of wild-type, TDP-43Q331K/Q331K, and TDP-43Q331K/+ mice.
(c) Automated continuous behavioural monitoring (ACBM) of 4-month-old mice (n = 10 mice per genotype; 5 males and 5 females). Significantly altered behaviours are displayed: walking: interaction P<0.0001; hanging: interaction P=0.002; rearing: interaction P=0.038; eating-by-hand: genotype P=0.008; repeated measures two-way ANOVA.
(d) Walking behaviour as assessed by ACBM in 7.5-month-old male and female mice (n = 5 mice per genotype). Walking male: interaction P<0.0001; walking female: interaction P=0.334; repeated measures two-way ANOVA.
(e) Ratios of mice genotyped at 10 days (all of which were successfully weaned) broken down by gender. Female (χ2=2.311, d.f.=2, P=0.315), Male (χ2=7.612, d.f.=2, P=0.022); Chi square test.
Error bars represent mean ± s.e.m.