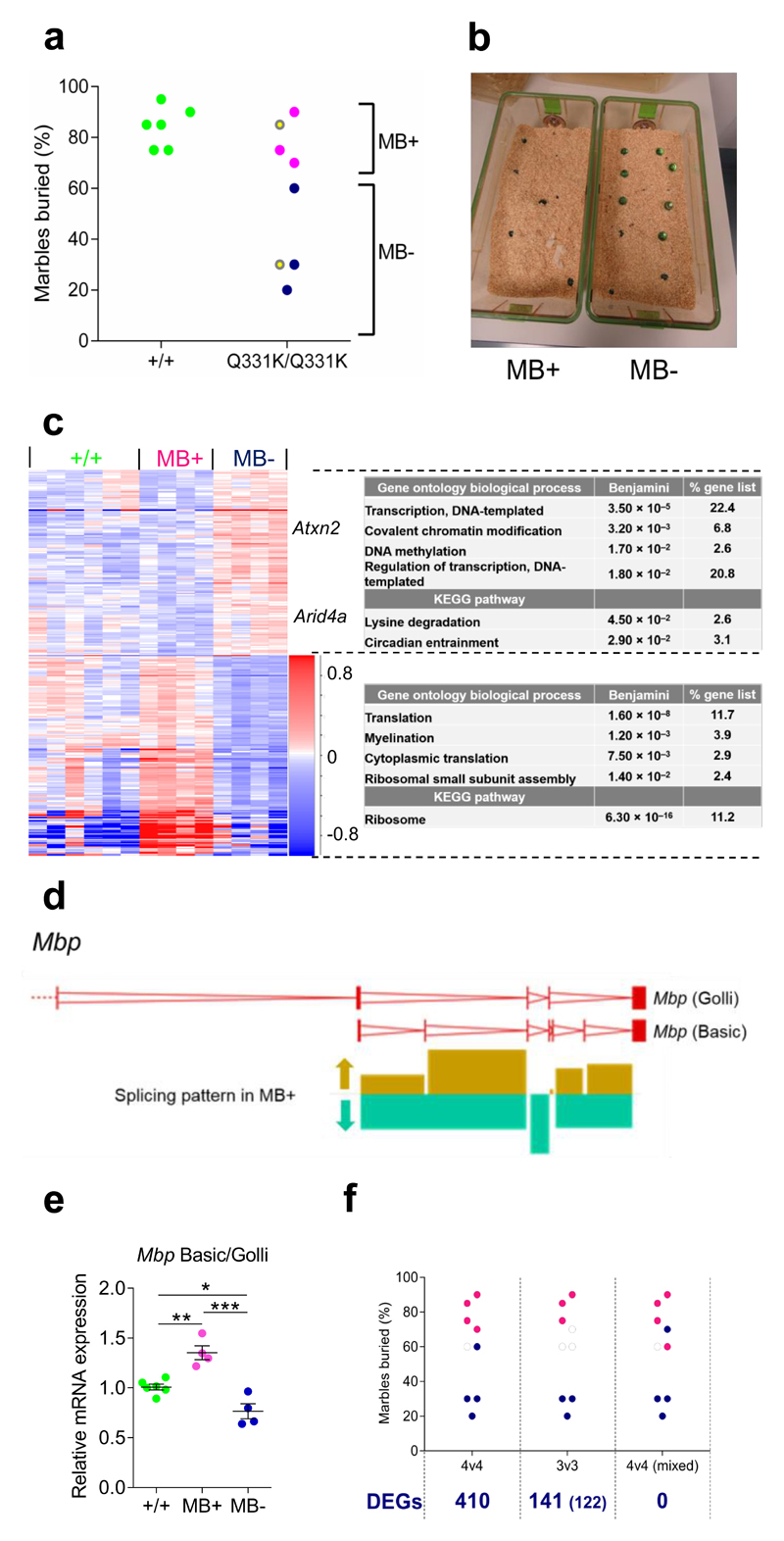
Figure 7. Phenotypic stratification of transcriptomic data from mutant mice allows the identification of putative disease modifiers.
(a) Marble-burying in 5-month-old mice prior to sacrifice. MB+ mice bury at or above the median number of marbles for the group, and MB- mice bury fewer. Yellow dots indicate TDP-43Q331K/Q331K littermates.
(b) Marble burying activity of TDP-43Q331K/Q331K littermates as described in (a).
(c) Hierarchical clustering of DEGs in frontal cortices comparing MB+ and MB- TDP-43Q331K/Q331K mice. Genes Atxn2 and Arid4a are highlighted (n = 6 wild-type, 4 MB+ TDP-43Q331K/Q331K and 4 MB- TDP-43Q331K/Q331K mice). Comparison: DESeq2 MB+ v MB-. Gene ontology (GO) biological processes and KEGG pathway enriched terms are displayed.
(d) Graphical representation of altered splicing of Mbp. Arrows indicate the altered pattern of splicing in MB+ relative to MB- TDP-43Q331K/Q331K mice.
(e) qPCR of the ratio of Mbp Basic to Mbp Golli (n = 6 wild-type, 4 TDP-43Q331K/+, 4 TDP-43Q331K/Q331K mice). Pairwise comparisons: wild-type vs. MB+: P=0.005 (**); wild-type vs. MB-: P=0.024 (*); MB+ vs. MB-: P=0.0003 (***); one-way ANOVA followed by Holm-Sidak post-hoc tests. Error bars denote s.e.m.
(f) Representative marble burying analyses: 4:4, original analysis; 3:3, comparing the three best MB+ and three worst MB- mice; 4v4 mixed, one MB- mouse swapped with one MB+ mouse. Number of DEGs identified by DESeq2 comparison of MB+ v MB- mice for each comparison is given below. For 3:3, hits common to the 4:4 stratification are shown in brackets.