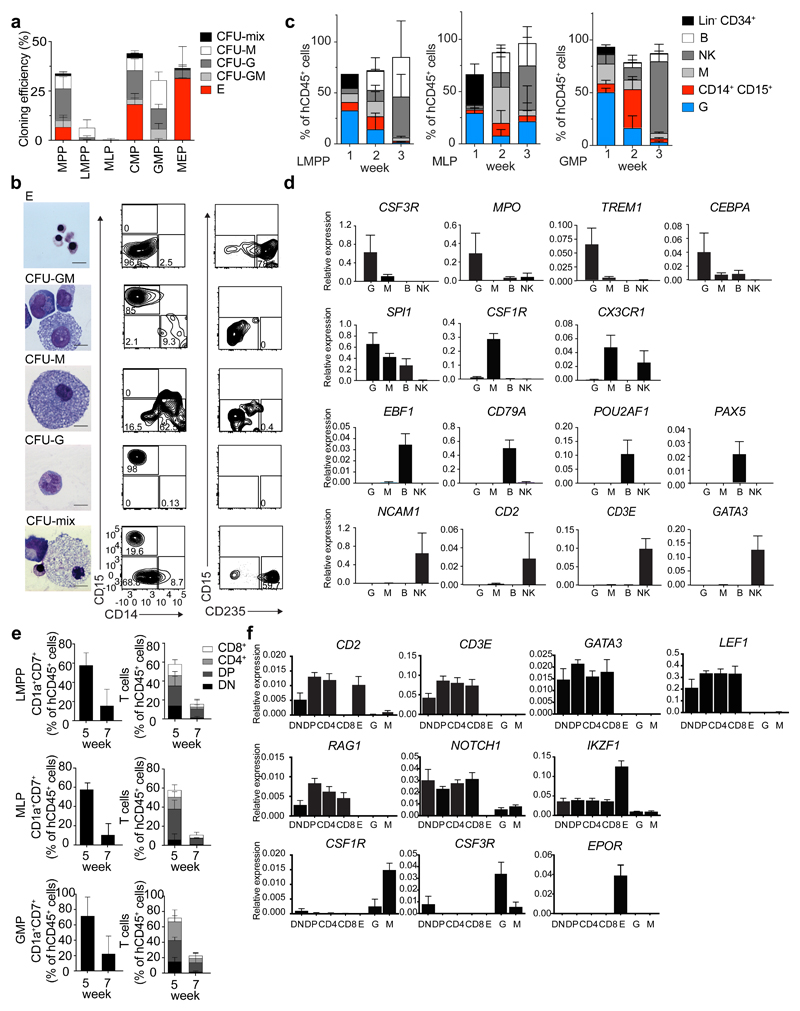
Figure 1. Human CB lympho-myeloid populations have distinct functional potential in vitro.
(a) Cloning efficiency and lineage affiliation of myelo-erythroid colonies in a CFU assay (150 CB HSPCs plated). Error bars are ± SD. n=5. CFU-mix, mixed erythro-myeloid colony; CFU-M, monocyte/macrophage colony; CFU-G, granulocyte colony; CFU-GM, granulocyte and monocyte/macrophage colony; E, erythroid colony (BFU-E and CFU-E). (b) Morphology of May-Grunwald-Giemsa stained cells from CFU assay (left, bar size 10 µm) and flow cytometric plots of cells harvested from indicated colony types (right). (c) Lineage output after culturing 150 LMPP, MLP and GMP cells for 1, 2 or 3 weeks on MS-5 stroma with SCF, G-CSF, FLT3L, IL15, IL2 and DuP-697. Data represent mean from 3 CB donors ± SD. Flow cytometric plots for two week cultures shown in Supplementary Fig. 1b. (d) Gene expression analysis of flow cytometric-purified output cells from (c). (e) T cell output after culturing LMPP, MLP and GMP cells in bulk for 5 or 7 weeks on OP9-hDL1 stroma with SCF, FLT3L and IL7. Data represent percentage from hCD45+ cells from 5 CB donors (mean± 1SD). DN, CD7+CD1a+ CD4-CD8-; DP, CD7+CD1a+CD4+CD8+; CD4, CD7+CD1a+CD4+CD8-; CD8, CD7+CD1a+CD4-CD8+. Flow cytometric plots for 5 week cultures shown in Supplementary Fig. 1c. (f) Gene expression analysis from flow-purified output cells from (e) and control mature non-T cells, obtained from sorting cells from E, G and M colonies.