Citrus CsMADS6 coordinately and positively modulates carotenoid metabolism by directly regulating the expression of LCYb1 and other carotenogenic genes, including PSY, PDS, and CCD1.
Abstract
Although remarkable progress has been made toward understanding carotenoid biosynthesis, the mechanisms that regulate the transcription of carotenogenic genes remain poorly understood. Lycopene β-cyclases (LCYb) are critical enzymes located at the branch point of the carotenoid biosynthetic pathway. Here, we used the promoter sequence of LCYb1 as bait in a yeast one-hybrid screen for promoter-binding proteins from sweet orange (Citrus sinensis). This screen identified a MADS transcription factor, CsMADS6, that was coordinately expressed with fruit development and coloration. Acting as a nucleus-localized transcriptional activator, CsMADS6 directly bound the promoter of LCYb1 and activated its expression. Overexpression of CsMADS6 in citrus calli increased carotenoid contents and induced the expression of LCYb1 and other carotenogenic genes, including phytoene synthase (PSY), phytoene desaturase (PDS), and carotenoid cleavage dioxygenase1 (CCD1). CsMADS6 up-regulated the expression of PSY, PDS, and CCD1 by directly binding to their promoters, which suggested the multitargeted regulation of carotenoid metabolism by CsMADS6. In addition, the ectopic expression of CsMADS6 in tomato (Solanum lycopersicum) affected carotenoid contents and the expression of carotenogenic genes. The sepals of CsMADS6-overexpressing tomato lines exhibited dramatic changes in carotenoid profiles, accompanied by changes in plastid ultrastructure. Global transcriptome analysis of transgenic sepals revealed that CsMADS6 regulates a series of pathways that promote increases in flux through the carotenoid pathway. Overall, these findings establish that CsMADS6 directly regulates LCYb1 and other carotenogenic genes to coordinately and positively modulate carotenoid metabolism in plants, which may provide strategies to improve the nutritional quality of crops.
Carotenoids are important secondary metabolites that are widely distributed and play many vital roles in nature. In plants, carotenoids generally give flowers and fruits distinct colors ranging from yellow and orange to red (Bartley and Scolnik, 1995). They also participate in photosynthesis and photoprotection (Niyogi et al., 1997; Holt et al., 2005), serve as precursors for phytohormones (Schwartz et al., 1997; Alder et al., 2012), and promote the scavenging of reactive oxygen species (Di Mascio et al., 1989; Edge et al., 1997). In humans, carotenoids serve as precursors for vitamin A (DellaPenna and Pogson, 2006) and excellent antioxidants that prevent cancer and other related diseases (Fraser and Bramley, 2004; Fiedor and Burda, 2014). The carotenoid biosynthesis pathway has been studied extensively, and genes underlying most of the biosynthetic steps have been identified in many plants (Fraser et al., 1994; Moise et al., 2014; Nisar et al., 2015). Previous studies have shown that the coordinated expression of genes that encode carotenoid biosynthetic enzymes tightly regulates carotenoid metabolism (Liu et al., 2015; Yuan et al., 2015). However, although the carotenoid biosynthetic pathway and the expression patterns of carotenogenic genes are well characterized, knowledge of the transcriptional regulatory mechanisms that control the expression of these genes is rather limited.
Transcription factors (TFs) bind to promoters and directly regulate the transcription of their target genes. A large number of TFs affect carotenoid accumulation by regulating fruit ripening, ethylene biosynthesis, photomorphogenesis, and other processes. These TFs include TAGL1 (TOMATO AGAMOUS-LIKE1; Itkin et al., 2009; Vrebalov et al., 2009), ERF6 (ETHYLENE RESPONSE FACTOR6; Lee et al., 2012), GLK2 (GOLDEN2-LIKE; Powell et al., 2012), and CubHLH1 (BASIC HELIX-LOOP-HELIX1; Endo et al., 2016). However, only a few TFs have been demonstrated to directly regulate the expression of carotenogenic genes and control carotenoid metabolism. For example, PIF1 (PHYTOCHROME INTERACTING FACTOR1), a light-responsive TF, specially binds the PSY (phytoene synthase) promoter and represses the expression of PSY and the accumulation of carotenoids in dark-grown Arabidopsis (Arabidopsis thaliana) seedlings (Toledo-Ortiz et al., 2010) and tomato (Solanum lycopersicum) fruits (Llorente et al., 2016). However, HY5 (LONG HYPOCOTYL5), a PIF antagonist, directly binds a common cis-element (G-box) in the promoter of PSY and promotes the accumulation of carotenoids associated with photosynthesis in response to light and temperature cues (Toledo-Ortiz et al., 2014). Welsch et al. (2007) identified the APETALA2 (AP2)/ethylene-response element-binding protein RAP2.2, which binds the ATCTA element in the promoter of PSY and regulates its expression in Arabidopsis. Tomato RIN (RIPENING INHIBITOR), a MADS-box TF, promotes the accumulation of lycopene by directly regulating the expression of PSY (Martel et al., 2011). Recently, papaya (Carica papaya) CpNAC1 was shown to bind directly the NAC-binding site in CpPDS2/4 (phytoene desaturase) promoters (Fu et al., 2016). CpNAC2 interacts with CpEIN3a and cooperatively regulates the expression of the carotenoid biosynthesis-related genes CpPDS2/4, CpLCYe (lycopene ε-cyclase), and CpCHY-b (β-carotene hydroxylase) during fruit ripening (Fu et al., 2017). Sweet osmanthus (Osmanthus fragrans) OfWRKY3 binds the W-box palindrome motif in the OfCCD4 (carotenoid cleavage dioxygenase4) promoter and induces gene expression (Han et al., 2016). Zhu et al. (2017) identified an R2R3-MYB TF, CrMYB68, that negatively regulates the expression of CrBCH2 (β-carotene hydroxylase) and CrNCED5 (9-cis-epoxycarotenoid dioxygenase) in the flavedo of Citrus reticulata.
MADS proteins are one of the largest TF families in plants (Shore and Sharrocks, 1995; Kaufmann et al., 2005). An increasing number of studies have shown that these proteins participate in the regulation of fruit ripening and the carotenoid biosynthesis that is associated with fruit ripening. For example, mutations in the tomato MADS-box genes RIN, TAGL1, and FUL1/2 (FRUITFULL1/2) result in fruit ripening-defective phenotypes with repressed carotenoid accumulation (Vrebalov et al., 2002, 2009; Itkin et al., 2009; Bemer et al., 2012; Wang et al., 2014). Overexpression of either tomato TAGL1 or its homolog PpPLENA from peach (Prunus persica) in tomato produces succulent sepals that accumulate substantial levels of lycopene (Itkin et al., 2009; Tadiello et al., 2009; Vrebalov et al., 2009). RNA interference-mediated suppression of the gene encoding SlMADS1, a negative regulator, enhances both the expression of PSY1 and the accumulation of carotenoids in tomato fruit (Dong et al., 2013). Most of the limited numbers of TFs that are known to affect carotenoid biosynthesis directly regulate PSY, which encodes the enzyme that catalyzes the first and rate-determining step of carotenoid biosynthesis. However, knowledge of TFs that coordinately modulate carotenoid metabolism by directly regulating other biosynthetic genes is still lacking, even in model plants.
Lycopene β-cyclases (LCYb) catalyze the cyclization of red lycopene into yellow-orange β-carotene at the branch point of the carotenoid biosynthesis pathway (Cunningham et al., 1996). The levels of LCYb expression change dramatically when carotenoids accumulate during fruit development in a range of plant species (Pecker et al., 1996; Yuan et al., 2015). Numerous studies have demonstrated that LCYb genes are transcriptionally regulated by many factors, such as phytohormones and ripening-associated TFs. For example, during tomato fruit ripening, the expression of LCYb genes is repressed by elevated levels of ethylene, leading to the massive accumulation of lycopene, which is responsible for the red color of ripe fruit (Fraser et al., 1994; Ronen et al., 2000; Alba et al., 2005). Repression of the MADS-box gene TAGL1 or the ethylene signaling gene SlAP2a up-regulates the expression of LCYb, which could account for the significantly decreased lycopene and increased β-carotene that is responsible for the orange-yellow phenotype of ripe fruit (Vrebalov et al., 2009; Chung et al., 2010). Similarly, silencing of the AUXIN RESPONSE FACTOR2, which displays marked ripening-associated expression, suppresses ethylene production and promotes the expression of SlLCYB and SlCYCB (chromoplast-specific lycopene β-cyclase; Hao et al., 2015). However, the mechanisms used by these ripening-associated regulators to regulate the expression of LCYb remain unknown.
Citrus is a nonclimacteric fruit type that accumulates high levels of carotenoids (Fanciullino et al., 2006; Xu et al., 2006; Matsumoto et al., 2007). In citrus fruits that predominantly accumulate β,β-xanthophylls (i.e. violaxanthin and β-cryptoxanthin), the transcriptional regulation of the LCYb genes during ripening has a remarkable influence on the carotenoid profile. High-level expression of LCYb genes shifts the flux of the pathway to the β-branch during the orange stage (Kato et al., 2004; Zhang et al., 2012; Rodrigo et al., 2013). Additionally, reductions in the transcript levels of a chromoplast-specific LCYb gene are responsible for the substantial accumulation of lycopene in the pulp of sweet orange (Citrus sinensis; Lu et al., 2016b) and grapefruit (Citrus paradisi; Alquézar et al., 2009; Mendes et al., 2011). However, whether and how the expression of the citrus LCYb genes is regulated by the ripening-associated regulators that are mentioned above are not clear. We previously identified two LCYb genes (LCYb1 and LCYb2) from sweet orange and successfully identified critical regulatory elements in their promoters (Lu et al., 2016a, 2016b). In this study, we used the promoter sequences of these LCYb genes as bait to screen a sweet orange fruit cDNA library and identified a MADS TF. Based on this cDNA sequence, this gene was identical to a gene that has been isolated previously from mandarin (Citrus unshiu) named CitMADS6 (Endo et al., 2006). However, whether and how CitMADS6 modulates carotenoid metabolism are currently unknown. Here, we identified CsMADS6 in sweet orange and found that it induced increases in carotenoid metabolism by directly regulating the expression of LCYb1 and other carotenogenic genes. Its roles in regulating the carotenoid metabolism of fruit were characterized subsequently in transgenic tomato. Finally, multiple regulatory functions of CsMADS6 in reprogramming transcriptional networks that support the biosynthesis and accumulation of carotenoids are discussed. These findings advance our understanding of the complex transcriptional regulation of carotenoid metabolism in plants.
RESULTS
Yeast One-Hybrid Screening and Sequence Analysis of CsMADS6
To identify TFs that regulate carotenoid metabolism, we performed a yeast one-hybrid (Y1H) screen. The promoter sequences of LCYb1 and LCYb2 were used as bait to screen a citrus fruit cDNA library. After Y1H screening, we successfully obtained several clones (Supplemental Table S1). One clone encoding a protein belonging to the large MADS TF superfamily attracted our attention because an increasing number of studies have demonstrated that MADS protein(s) play important roles in fruit development (Giovannoni, 2004). A BLAST search of the National Center for Biotechnology Information (NCBI) database revealed that this MADS sequence was identical to a previously isolated gene, CitMADS6, from mandarin (Endo et al., 2006). Thus, we named this apparently orthologous gene from sweet orange CsMADS6. We successfully obtained its full-length \coding sequence, which encoded a protein of 257 amino acids with a calculated molecular mass of 29.64 kD and a predicted pI of 9.37.
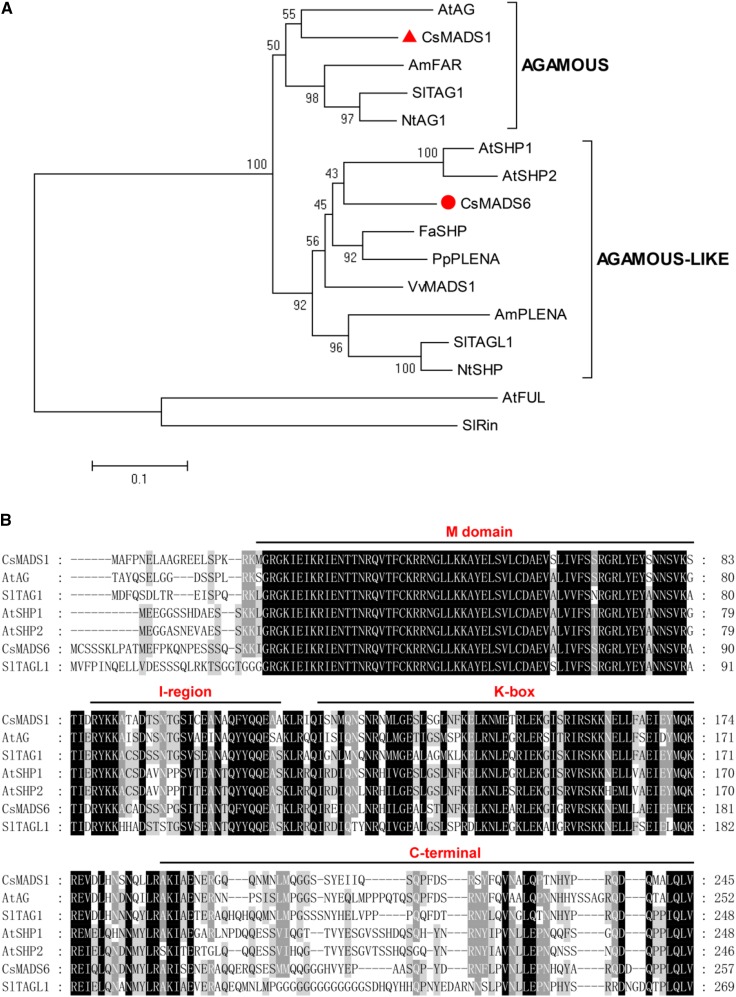
To understand the relationship of this CsMADS6 protein to other MADS proteins, we generated a phylogenetic tree with CsMADS6 and other C-type MADS proteins. A paralogous gene, CsMADS1 (corresponding to the CitMADS1 from mandarin), encoding a protein with 69% amino acid sequence similarity to CsMADS6, also was included in this analysis. The analysis indicated that CsMADS6 and CsMADS1 grouped with the AGAMOUS-like and AGAMOUS subclades, respectively. CsMADS6 was closely linked with Arabidopsis AtSHP2 and tomato TAGL1 (Fig. 1A). A multiple sequence alignment of CsMADS6, CsMADS1, and their homologs from Arabidopsis and tomato indicated that these proteins contained four typical plant MADS regions (the M-domain, I-region, K-box, and C-terminal domain). The sequences from the conserved M-domain were highly similar, followed by the less similar sequences from the I-region and the K-box. The C-terminal domain generally varied (Fig. 1B).
Figure 1.
Sequence analysis of CsMADS6. A, Phylogenetic analysis of CsMADS6 and related proteins from other plant species. The scale bar represents 0.1 substitutions per site. The red circle and triangle indicate CsMADS6 and CsMADS1, respectively. Arabidopsis AtFUL and tomato SlRIN were used as outgroups. B, Multiple sequence alignment of CsMADS6 and related proteins from Arabidopsis and tomato. The horizontal lines mark the four conserved domains.
Expression of CsMADS6 Increases with Fruit Development and Coloration
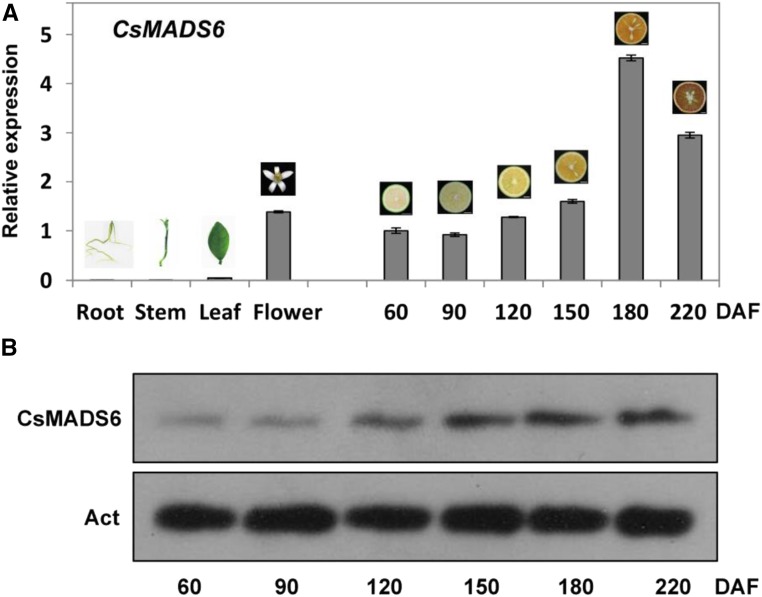
To understand the spatial and temporal expression patterns of CsMADS6, we determined its relative transcript levels in different tissues and during the different stages of fruit development in sweet orange. CsMADS6 was expressed predominantly in the flower and fruit tissues. We observed low levels of expression in roots, stems, and leaves. During fruit development, the transcript levels of CsMADS6 increased gradually, peaking at 180 d after flowering (DAF; breaker stage) and declining at 220 DAF (senescence stage; Fig. 2A).
Figure 2.
Transcript and protein levels of CsMADS6 in citrus. A, Relative expression levels of CsMADS6 in different tissues and at different stages of fruit ripening. The data are expressed as means ± sd of three replications. B, Protein levels of CsMADS6 at different stages of fruit ripening. Act, ACTIN protein (internal reference).
We used immunoblotting to determine the levels of the CsMADS6 protein in citrus fruits at different stages of ripening. ACTIN was used as a loading control. The protein levels of CsMADS6 increased gradually during fruit development (Fig. 2B). This result agreed well with the analysis of mRNA levels. Based on these data, we concluded that the expression of CsMADS6 increased with the coloration of citrus fruit and the accumulation of carotenoids (Liu et al., 2007).
CsMADS6 Localizes to the Nucleus and Functions as a Transcriptional Activator
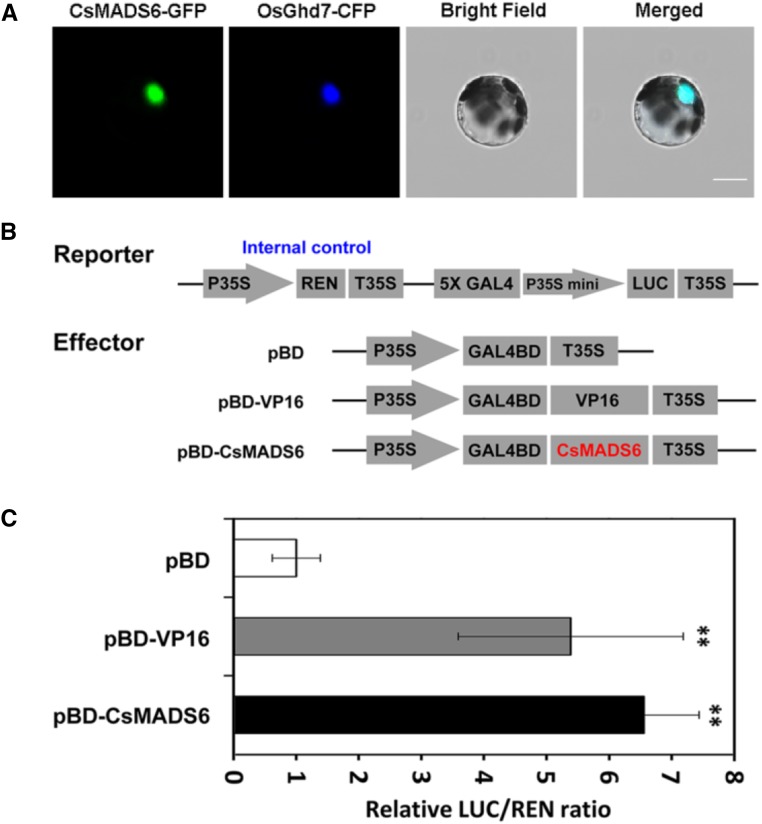
To test the subcellular localization of CsMADS6, we generated a CsMADS6-GFP fusion construct and coexpressed it with the nuclear marker OsGhd7-CFP (CYAN FLUORESCENT PROTEIN) in citrus protoplasts. Using fluorescence microscopy, we found that the GFP signals overlapped extensively with the CFP signals, indicating that the CsMADS6-GFP fusion protein colocalized with the OsGhd7-CFP nuclear marker in vivo (Fig. 3A). These data indicated that the CsMADS6 protein localized to the nucleus.
Figure 3.
Subcellular localization and transcriptional activity of CsMADS6. A, Subcellular localization of CsMADS6 in citrus protoplasts. CsMADS6-GFP was cotransformed with OsGhd7-CFP, which was used as a nuclear marker. CsMADS6-GFP, GFP signal; OsGhd7-CFP, CFP signal; Bright Field, white light; Merged, combined GFP and CFP signals. Bar = 10 μm. B, Schematic diagrams of vectors used for the transcriptional activity assay. pBD-CsMADS6, Vector containing CsMADS6; pBD and pBD-VP16 served as the negative and positive controls, respectively. C, Transcriptional activity of CsMADS6. The transcriptional activity of CsMADS6 was quantified using a luciferase assay. The data are expressed as means ± sd from six biological replicates. Asterisks indicate significant differences relative to the empty vector control by one-way ANOVA tests (**, P < 0.01).
To assess the transcriptional activity of CsMADS6 in vivo, we used a transient expression assay system in Nicotiana benthamiana leaves. The reporter vector contained 5× GAL4 activation domains upstream of the LUC (Firefly luciferase) gene. In the effector vector, the CsMADS6 gene was fused to the GAL4 DNA-binding domain (pBD-CsMADS6). Transactivation of the LUC gene was measured relative to the REN (Renilla luciferase) gene under the control of the constitutive Cauliflower mosaic virus 35S (CaMV35S) promoter (Fig. 3B). These constructs were coexpressed in N. benthamiana leaves, and luciferase activity was quantified. The results showed that, compared with the negative control (pBD), luciferase activity increased significantly when the LUC-containing construct was coexpressed with pBD-CsMADS6, reaching levels similar to those of the positive control (pBD-VP16; Fig. 3C). These data provided evidence that CsMADS6 may function as a transcriptional activator.
CsMADS6 Directly Binds and Activates the Promoter of LCYb1
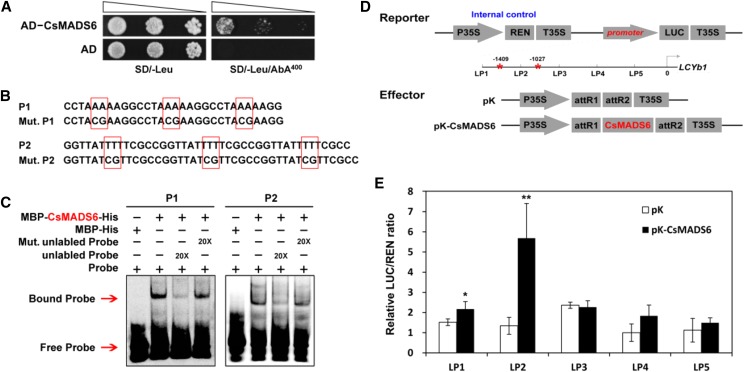
To test the interaction between the CsMADS6 protein and the LCYb1 promoter, we first performed a Y1H assay. Bait yeast cells cotransformed with the control vector (activation domain [AD]) or the fusion vector (AD-CsAMDS6) grew well on synthetic dropout medium (SD) without Leu. However, only the yeast cells cotransformed with the fusion vector AD-CsMADS6 could survive on the selective medium supplemented with 400 ng mL−1 aureobasidin A (AbA; Fig. 4A). These data indicated that the CsMADS6 protein interacted with the LCYb1 promoter in the yeast system.
Figure 4.
Interaction of the CsMADS6 protein with the promoter of LCYb1. A, Y1H assay showing the binding of CsMADS6 to the LCYb1 promoter. AD, Empty vector used as the negative control; AD-CsMADS6, prey vector containing CsMADS6; SD/-Leu, SD medium without Leu; SD/-Leu/AbA400, SD medium without Leu supplemented with AbA at the concentration of 400 ng mL−1. Transformed yeast cells were dotted at 10−1 dilutions on the selective medium. B, Probes used for EMSA. P1 and P2, Two Cy5-labeled probes containing three copies of the CArG elements present in the LCYb1 promoter; Mut, P1 and P2 mutant probes (the mutated sites are surrounded by red boxes). C, EMSA showing the binding of CsMADS6 to the LCYb1 promoter. + and − indicate the presence and absence of the indicated probe or protein; 20× indicates a 20-fold excess of unlabeled probe or mutant unlabeled probe. Red arrows indicate the positions of protein-DNA complexes or free probes. D, Schematic diagrams of vectors used for the dual-luciferase assay. The reporter vector contained different promoter fragments of LCYb1 fused to LUC. Red asterisks indicate the positions of the two CArG elements. pK, Empty vector; pK-CsMADS6, overexpression vector containing CsMADS6. E, Dual-luciferase assay showing relative CsMADS6 activation of different promoter fragments of LCYb1. The data are expressed as means ± sd from at least four biological replicates. Asterisks indicate significant differences relative to the empty vector control by one-way ANOVA tests (*, P < 0.05 and **, P < 0.01).
Plant MADS-box proteins bind to specific DNA sequences known as CArG elements [CArG element sequences: C(C/T)(A/T)6(A/G)G, C(A/T)8G and C(C/T)(A/T)G(A/T)4(A/G)G; Ito et al., 2008; Fujisawa et al., 2011]. We found two possible CArG elements in the LCYb1 promoter (Fig. 4B; Supplemental Table S2). We performed an electrophoretic mobility shift assay (EMSA) to test whether the CsMADS6 protein could bind the LCYb1 promoter at these potential binding sites. Recombinant MBP-CsMADS6-His shifted a band with the labeled DNA probes (P1 or P2), presumably by generating a protein-DNA complex. In contrast, the MBP-His control did not affect the mobility of the labeled DNA probes. After an excess of unlabeled DNA probe was added, the shifted band completely disappeared. In contrast, when an excess of mutated unlabeled DNA probe was added, MBP-CsMADS6-His still reduced the mobility of the labeled DNA probe (Fig. 4C). We obtained similar results with two different DNA probes from the LCYb1 promoter, indicating that the CsMADS6 protein specifically bound each CArG element in the LCYb1 promoter.
Thus, we found that CsMADS6 directly bound to the LCYb1 promoter in vivo and in vitro. We used a dual-luciferase assay to investigate the effect of CsMADS6 binding on promoter activity. The full-length promoter of LCYb1 was divided into five fragments (LP1–LP5), relative to the two CArG elements, by successively deleting sequences from the 5′ end. The promoter activity of each fragment was measured to identify the region(s) that interacted with CsMADS6 (Fig. 4D). As shown in Figure 4E, the relative luciferase expression driven by the promoter fragment of LP1 was significantly higher in the presence of CsMADS6 than the control, suggesting that CsMADS6 activated the promoter activity of LP1. A deletion from LP1 to LP2 led to a high level of activation by CsMADS6, whereas a further deletion to LP3, LP4, or LP5 actually diminished activation. Taken together, the above data indicated that the CsMADS6 protein activated the promoter activity of LCYb1 by interacting with the two CArG elements that are located in the LP1 and LP2 fragments.
CsMADS6 Increases Carotenoid Biosynthesis and Affects the Expression of Carotenoid-Associated Genes
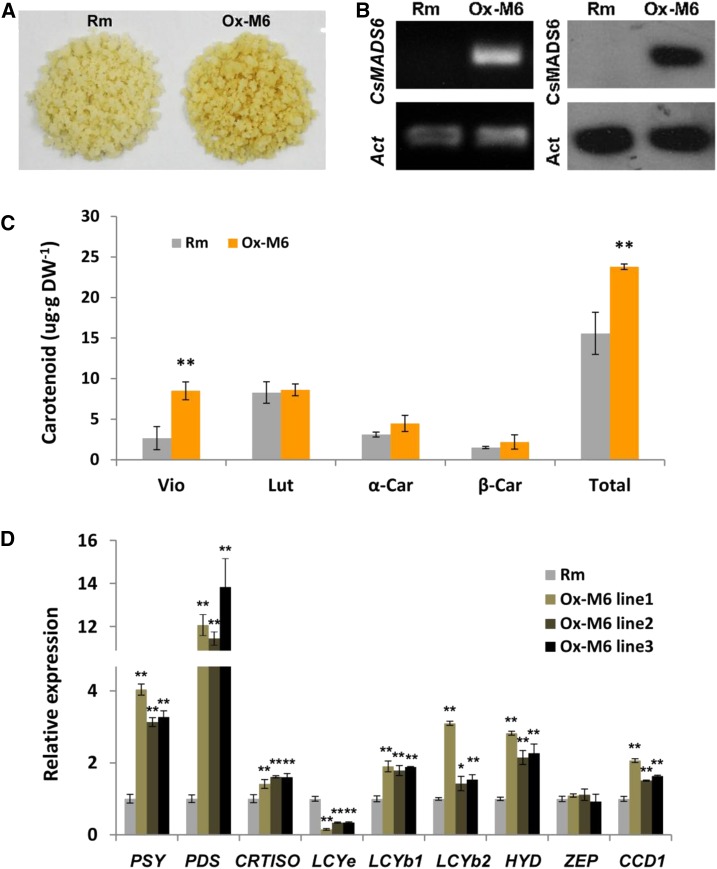
CsMADS6 was first transformed into citrus calli to examine whether it modulates carotenoid metabolism. Positive transformants from several independent lines were recovered. Most of the transgenic lines (Ox-M6) were slightly more yellow than the control during culture. We selected one representative transgenic line that showed color change and accumulated high levels of CsMADS6 transcript and protein for further carotenoid analysis (Fig. 5, A and B). Violaxanthin, lutein, α-carotene, and β-carotene were identified as the predominant carotenoids in these citrus calli and were quantified using HPLC. The concentrations of the other carotenoids were below the detection limit. The levels of violaxanthin increased significantly, contributing to the high levels of total carotenoids in the CsMADS6-overexpressing calli (Fig. 5C).
Figure 5.
Effects of CsMADS6 overexpression in citrus calli. A, Phenotypes of transgenic citrus calli. Rm, The wild type; Ox-M6, CsMADS6-overexpressing citrus calli. B, Reverse transcription (RT)-PCR and immunoblotting analysis of CsMADS6 transcript and protein levels. Act, ACTIN gene (internal reference). C, Carotenoid content in transgenic citrus calli. Vio, Violaxanthin; Lut, lutein; α-Car, α-carotene; β-Car, β-carotene; Total, total carotenoids; DW, dry weight. D, Relative expression levels of carotenoid biosynthetic genes in transgenic citrus calli. The name of the pertinent gene is indicated for each set of bars. Data are represented as means ± sd of three replicates. Asterisks indicate significant differences relative to the control by one-way ANOVA tests (*, P < 0.05 and **, P < 0.01).
The transcript levels of carotenoid biosynthetic genes were monitored in three independent transgenic lines. Notably, LCYb1 expression was induced significantly in the overexpressing lines, consistent with CsMADS6 up-regulating the transcription of LCYb1. We analyzed the expression of other important biosynthetic genes, including PSY, PDS, CRTISO (carotenoid isomerase), LCYe, LCYb2, HYD (a/β-carotene hydroxylase), ZEP (zeaxanthin epoxidase), and CCD1. Using reverse transcription quantitative PCR (RT-qPCR), we observed a marked increase in the expression of PSY, PDS, CRTISO, LCYb2, HYD, and CCD1 in the CsMADS6-overexpressing lines. The transcript levels of PSY, PDS, and CCD1 increased 4-, 12-, and 2-fold in Ox-M6 lines, respectively. In contrast, the transcript levels of LCYe were repressed and ZEP expression did not change significantly in Ox-M6 lines (Fig. 5D). Taken together, these results indicated that the overexpression of CsMADS6 increased carotenoid biosynthesis in transgenic calli and induced the expression of LCYb1 and other carotenogenic genes, including PSY, PDS, and CCD1.
We also quantified the expression of TFs that are known to play a regulatory role in carotenoid metabolism, such as HY5, PIF1, and RAP2.2 (Welsch et al., 2007; Toledo-Ortiz et al., 2010, 2014), in transgenic citrus calli. We found that the expression of HY5 and RAP2.2 increased significantly and that the expression of PIF1 was reduced notably in the CsMADS6-overexpressing lines (Supplemental Fig. S1). Moreover, RIN and FUL, two major ripening-associated MADS TF genes, were not expressed in citrus calli and were not expressed at significantly different levels in the transgenic lines and the control (data not shown).
CsMADS6 Directly Binds and Activates the Promoters of PSY, PDS, and CCD1
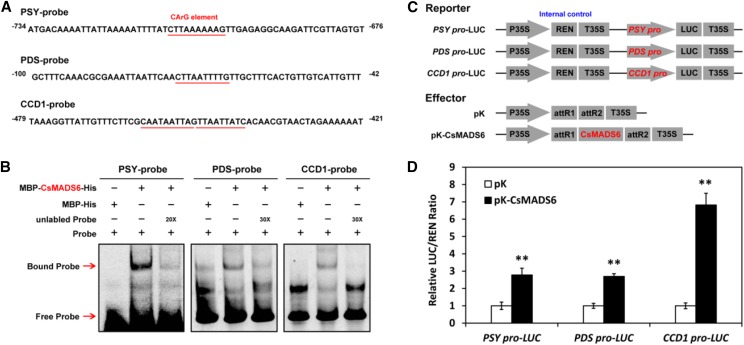
Because the transcript levels of carotenoid biosynthetic genes other than LCYb1, such as PSY, PDS, and CCD1, were induced significantly when CsMADS6 was overexpressed in citrus calli, we decided to investigate whether these genes were direct targets of CsMADS6. We first cloned the promoter regions of PSY (1,589 bp of accession no. MG594039), PDS (987 bp of accession no. MG594040), and CCD1 (1,295 bp of accession no. MG594041) from sweet orange and discovered at least one putative CArG element in each of their promoter regions (Fig. 6A; Supplemental Table S2). EMSAs were then performed to test whether the CsMADS6 protein binds these promoters. Due to the incomplete annealing of long probe sequences, we observed bands representing the free probes of PDS and CCD1 that did not influence the binding of CsMADS6 to the promoter fragments. When the labeled probes were incubated with the purified CsMADS6 protein, a specific shifted band was detected for each probe. After an excess of unlabeled probe was added, the shifted bands disappeared, suggesting that CsMADS6 bound specifically to these DNA fragments (Fig. 6B).
Figure 6.
Interaction of the CsMADS6 protein with the promoters of PSY, PDS, and CCD1. A, Probes used for EMSA. The predicted CArG elements are underlined in red. Numbers indicate the positions relative to the ATG start codon. B, EMSA showing the binding of CsMADS6 to the promoters of PSY, PDS, and CCD1. + and − indicate the presence and absence, respectively, of a probe or protein; 20× and 30× indicate 20-fold and 30-fold excess, respectively, of unlabeled probes. Red arrows indicate the positions of protein-DNA complexes or free probes. C, Schematic diagrams of vectors used for the dual-luciferase assay. In the reporter vectors, the promoters from PSY, PDS, and CCD1 are fused to the LUC reporter gene. pK, Empty vector; pK-CsMADS6, CsMADS6 overexpression vector. D, Dual-luciferase assay showing relative CsMADS6 activation of the PSY, PDS, and CCD1 promoters. The data are expressed as means ± sd from at least four biological replicates. Asterisks indicate significant differences relative to the empty vector control by one-way ANOVA tests (**, P < 0.01).
We used a dual-luciferase assay to evaluate the interactions between the CsMADS6 protein and the promoters of PSY, PDS, and CCD1 in vivo. The promoter sequences of PSY, PDS, and CCD1 were fused successfully to the LUC reporter gene (Fig. 6C). As shown in Figure 6D, CsMADS6 strongly activated these three promoters, especially the CCD1 promoter (approximately 7-fold). Thus, in addition to activating the promoter of LCYb1, CsMADS6 also directly bound and activated the promoters of PSY, PDS, and CCD1.
CsMADS6 Affects Carotenoid Profiles, Plastid Ultrastructure, and Carotenogenic Gene Expression in Transgenic Tomato
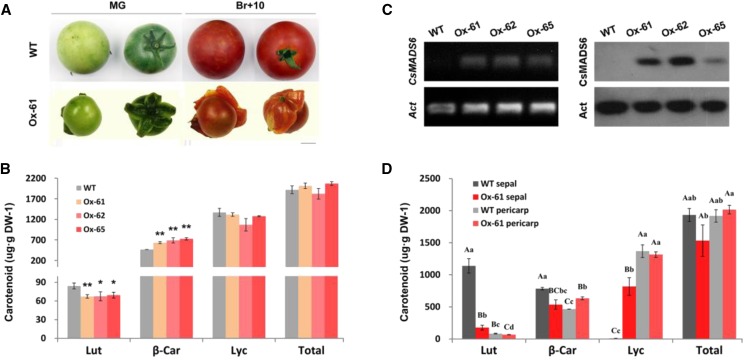
To better understand the function of CsMADS6 in the carotenoid metabolism of fruit, we ectopically overexpressed it in tomato, which is an optimal model system for this study because tomato is easily transformed and can produce fleshy fruit within a relatively short life cycle. Phenotypic characterization revealed that the transgenic sepals were fused, succulent, and exhibited the characteristics of fruit development, with the color changing from green to orange and, ultimately, to red (Fig. 7A). Three independent single-copy transgenic lines (Ox-61, Ox-62, and Ox-65) that accumulated high levels of CsMADS6 transcript and protein were chosen for further investigation (Fig. 7B).
Figure 7.
Effects of CsMADS6 overexpression in tomato fruits. A, Phenotypes of transgenic tomato fruits. WT, The wild type; Ox-61, an independent CsMADS6-overexpressing line; MG, mature green; Br+10, 10 d after breaker stage. Note the succulent sepals in the transgenic lines. Bar = 1 cm. B, RT-PCR and immunoblotting analysis of CsMADS6 expression. Act, ACTIN gene (internal reference). C and D, Carotenoid content in transgenic tomato pericarps (C) and sepals (D) at the Br+5 stage of fruit development. Lut, Lutein; β-Car, β-carotene; Lyc, lycopene; Total, total carotenoids; DW, dry weight. The data are expressed as means ± sd from three biological replicates. Statistically significant differences are indicated with either single asterisks or lowercase letters (P < 0.05) and double asterisks or uppercase letters (P < 0.01) by one-way ANOVA tests. Different letters within each column indicate significant differences.
The major carotenoid in ripening tomato fruits is lycopene, followed by β-carotene and then lutein (Ronen et al., 2000; Burns et al., 2003). Tomato fruit pericarps from the CsMADS6-overexpressing lines exhibited decreased levels of lutein, increased levels of β-carotene, and no changes in the levels of lycopene and total carotenoids relative to the control (Fig. 7C). In addition, we compared the carotenoid profiles of the sepals from the CsMADS6-overexpressing lines with that the wild type. Lutein is a photosynthetic pigment that accounts for the major fraction of carotenoids in the tomato sepal. In contrast to significantly reduced lutein and β-carotene content, lycopene levels in transgenic sepals increased dramatically to much higher levels than the trace amounts found in the wild-type sepals. Indeed, the carotenoid profiles of the transgenic sepals were similar to those of the pericarps from the wild type or the transgenic lines (Fig. 7D). Chlorophylls, which typically accumulate to high levels in wild-type sepals, were reduced to trace levels in the CsMADS6-overexpressing sepals (Supplemental Fig. S2). Based on these data, we concluded that the increased lycopene and decreased chlorophyll contents were responsible for the red appearance of the transgenic sepals, which had carotenoid profiles similar to those of the fruit pericarps.
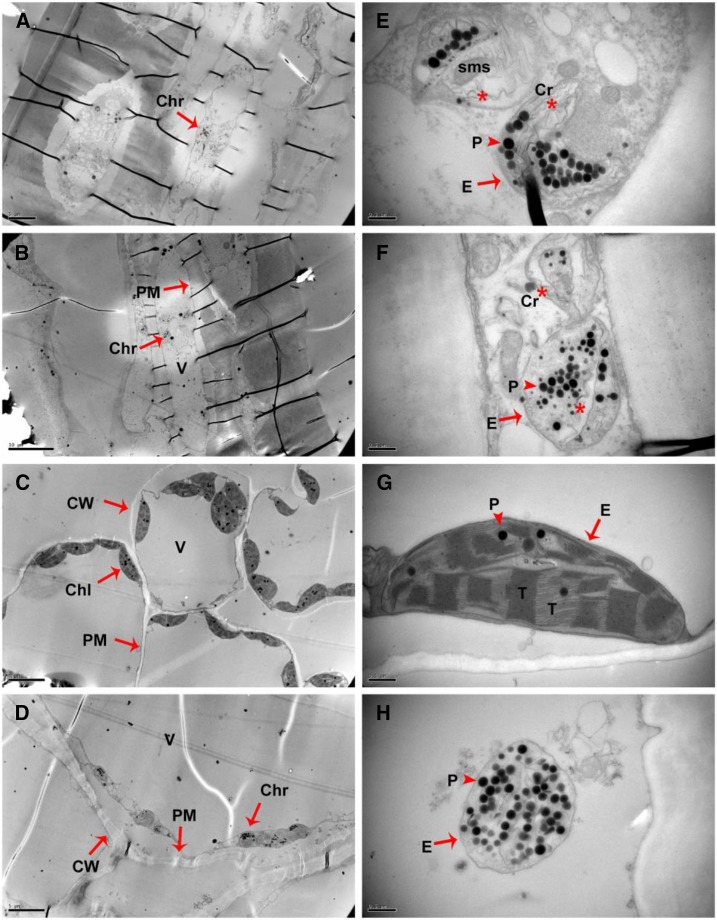
Carotenoids are stored in plastids, especially in chromoplasts (Vishnevetsky et al., 1999; Li and Yuan, 2013). We used transmission electron microscopy (TEM) to observe the plastid ultrastructure of transgenic tomato fruit. No significant differences were discovered in the ultrastructure of plastids from the pericarps of wild-type (Fig. 8, A and E) and transgenic (Fig. 8, B and F) tomato fruit. However, remarkable changes in plastid ultrastructure were observed in the sepals of the transgenic plants overexpressing CsMADS6. Instead of the spindle-shaped chloroplasts found in the wild-type sepals (Fig. 8, C and G), the plastids in the sepals of CsMADS6-overexpressing lines were small, round, contained numerous plastoglobuli (Fig. 8, D and H), and thus resembled chromoplasts from the pericarps of the wild type and the transgenic lines. This type of plastid ultrastructure was consistent with the accumulation of red lycopene to high levels in the sepals of CsMADS6-overexpressing lines.
Figure 8.
TEM of plastid ultrastructures from transgenic tomato fruits. A and E, Wild-type pericarps showing disrupted plastid envelopes. B and F, Transgenic pericarps exhibiting disorganized membrane systems. C and G, Wild-type sepals showing spindle-shaped plastids with many thylakoid plexuses and few plastoglobuli. D and H, Transgenic sepals with globular plastids with numerous plastoglobuli and intact envelopes. E to H are the magnified views of A to D, respectively. Abbreviations and symbols are as follows: Chl, chloroplast (arrow); Chr, chromoplast (arrow); Cr, carotenoid crystal remnant (asterisk); CW, cell wall (arrow); E, plastid envelope (arrow); P, plastoglobuli (arrowhead); PM, plasma membrane; sms, special membrane structure; T, thylakoid plexus; V, vacuole. Bars in A, C, and D = 5 μm; bar in B = 10 μm; and bars in E to H = 0.5 μm.
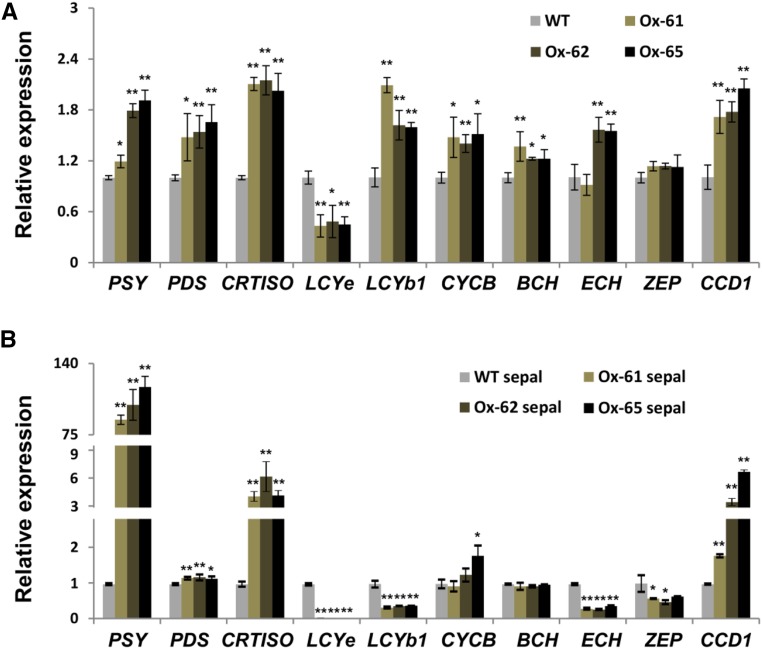
The expression of carotenoid biosynthetic genes in wild-type and transgenic tomato fruit pericarps was further examined by RT-qPCR. The results showed that in the CsMADS6-overexpressing lines, the expression levels of LCYb1, PSY, PDS, CRTISO, CYCB (the second functional LCYb gene in tomato fruits), BCH, ECH (α-carotene hydroxylase), and CCD1 were all significantly up-regulated relative to the wild type. However, the mRNA levels of LCYe were repressed and the expression of ZEP was not altered significantly in the transgenic lines (Fig. 9A). We also determined the transcript levels of LCYb1 and other carotenoid biosynthetic genes in the transgenic sepals of CsMADS6. We found that the mRNA levels of PSY in the transgenic sepals were approximately 100-fold greater than in the wild type (Fig. 9B). The mRNA levels of both CRTISO and CCD1 increased sharply, and the mRNA levels of PDS also were elevated in the transgenic sepals. In contrast, the expression of LCYe, LCYb1, ECH, and ZEP was significantly down-regulated in the transgenic sepals. The transcript levels of CYCB and BCH were not significantly different from those of their controls. The above analyses indicated that CsMADS6 affected carotenoid profiles and carotenogenic gene expression in transgenic tomato pericarps and sepals and also significantly promoted plastid development and chromoplast formation in transgenic tomato sepals.
Figure 9.
Relative expression levels of carotenoid biosynthetic genes in transgenic tomato. Gene expression from tomato pericarps (A) and sepals (B) at the Br+5 stage of fruit development is shown. WT, The wild type; Ox-61, Ox-62, and Ox-65, three independent CsMADS6-overexpressing lines. The name of the pertinent gene is indicated for each set of bars. Data are represented as means ± sd from three replicates. Asterisks indicate significant differences relative to the control by one-way ANOVA tests (*, P < 0.05 and **, P < 0.01).
Global Transcriptome Changes Contribute to the Dramatic Carotenoid Changes in Transgenic Tomato Sepals
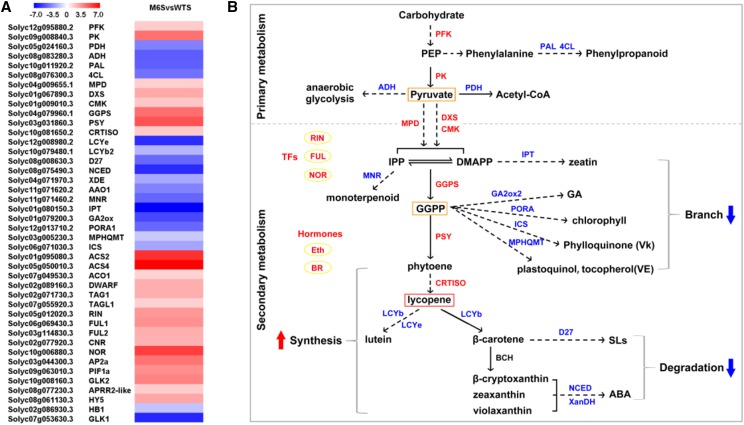
Since the carotenoid profiles and carotenogenic gene expression in transgenic tomato sepals changed dramatically, we performed RNA sequencing (RNA-seq) analysis to investigate the global transcriptome changes caused by CsMADS6 overexpression. The transcriptome of the transgenic sepals was compared with that of the wild-type sepals (M6SvsWTS) and the wild type pericarps (M6SvsWTR). A total of 4,435 differentially expressed genes (DEGs) were found in the M6SvsWTS comparison. A total of 2,983 DEGs were identified in the M6SvsWTR comparison (Supplemental Fig. S3). A full list of DEGs can be found in Supplemental Table S3. A correlation analysis revealed a high degree of consistency between the transcript abundance determined by RT-qPCR and RNA-seq (Supplemental Fig. S4). Hierarchical clustering analysis indicated that the expression patterns were classified into seven groups, and the expression profiles in M6S were more similar to those of WTR than to those of WTS (Supplemental Fig. S5). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses showed that the DEGs from the M6SvsWTS comparison were largely involved in metabolism and that the DEGs from the M6SvsWTR comparison were principally associated with photosynthesis (Supplemental Fig. S6). For subsequent analysis, we focused mainly on the DEGs that were identified from the M6SvsWTS comparison. A short list of DEGs, which is discussed below, is provided in Supplemental Table S4. The expression changes are provided in Figure 10A. Transcriptional regulation by the overexpression of CsMADS6 at a global level is schematically represented in Figure 10B.
Figure 10.
Transcriptome effects induced by CsMADS6 overexpression in transgenic tomato sepals. A, Heat map of selected carotenoid-associated DEGs from the M6SvsWTS comparison. The column represents the comparison of M6S (CsMADS6-overexpressing sepals) with WTS (wild-type sepals), and each row represents an individual gene. The intensity of log2 (fold change) is visualized using colors: red, up-regulated; blue, down-regulated. Gene identifiers and names are indicated at the left of each row. B, Schematic of the global transcriptional regulation by CsMADS6. Solid or dashed arrows indicate direct or indirect reaction flows in the pathway, respectively. The carotenoid-associated DEGs encoding enzymes that catalyze the reaction are shown at the sides of the arrows. Red and blue text denote expression levels with significant increases and decreases (P < 0.05), respectively. Yellow boxes highlight the critical intermediates in the metabolic pathways. The thick red and blue arrows indicate up- and down-regulation of the metabolic flux, respectively. Other processes, including primary pathways, secondary pathways, and phytohormone and TF levels (highlighted with yellow ovals), and their relation to accumulation of large amounts of a specific carotenoid (lycopene) in the transgenic sepals, are indicated. ABA, Abscisic acid; SLs, strigolactones.
In accordance with the preceding RT-qPCR analysis, the RNA-seq data revealed significantly high levels of PSY and CRTISO transcripts and low levels of LCYe transcripts in transgenic sepals. The expression levels of LCYb1 (Solyc04g040190) were not found due to the method that we used to quantify the RNA-seq data. The nucleotide sequence of LCYb2 (Solyc10g079480) shares 85% similarity with LCYb1, and the expression levels of LCYb2 were significantly down-regulated in transgenic sepals as determined by RNA-seq. Moreover, the expression levels of several late carotenoid metabolic genes that are involved in the enzymatic degradation of carotenoids for the production of abscisic acid and strigolactones, such as NCED and D27 (β-carotene isomerase D27), were reduced greatly in the transgenic sepals. The expression of genes involved in pathways other than the carotenoid biosynthetic pathway also was affected significantly by the overexpression of CsMADS6. For example, two predominant genes, PFK (6-phosphofructokinase) and PK (pyruvate kinase), which encode enzymes involved in carbohydrate metabolisms and that produce pyruvate, were up-regulated significantly in the M6SvsWTS comparison. Pyruvate dehydrogenase, which encodes an enzyme that catalyzes the decarboxylation of pyruvate to eventually produce acetyl-CoA for the citrate cycle (tricarboxylic acid cycle), was down-regulated significantly. Low transcript levels of alcohol dehydrogenase, which is involved in anaerobic respiration, were observed. Moreover, the transcript levels of many genes involved in secondary metabolic pathways other than the carotenoid biosynthetic pathway changed notably in the M6SvsWTS comparison. For example, the transcript abundances of two phenylpropanoid-related genes, Phe ammonia lyase and 4-coumarate-CoA ligase, were repressed substantially. The mevalonate (MVA) and methylerythritol 4-phosphate (MEP) pathways provide necessary precursors (isopentenyl pyrophosphate [IPP], dimethylallyl diphosphate [DMAPP], and geranylgeranyl pyrophosphate [GGPP]) for the biosynthesis of many secondary metabolites, including carotenoids, chlorophyll, plastoquinone, and tocopherol. The transcript levels of key genes, including mevalonate disphosphate decarboxylase, 1-d-deoxyxylulose 5-phosphate synthase, 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase, and geranylgeranyl pyrophosphate synthase, which encode enzymes involved in the biosynthesis of IPP, DMAPP, and GGPP, were increased significantly.
We also determined that the expression of genes associated with many secondary metabolic pathways that are located downstream of the MVA and MEP pathways were repressed significantly. For example, the transcript levels of the genes encoding IPT (tRNA dimethylallyl transferase), which is involved in catalyzing the biosynthesis of DMAPP into zeatin, and MNR (neomenthol dehydrogenase), which is involved in catalyzing the conversion of IPP into monoterpenoid, were both reduced. Additionally, genes that encode enzymes that are critical for utilizing GGPP for the synthesis of other metabolites also were down-regulated significantly in transgenic sepals; these genes include gibberellin 2-oxidase (responsible for GA biosynthesis), protochlorophyllide oxidoreductase (chlorophyll biosynthesis), isochorismate synthase (phylloquinone biosynthesis), and methyl-6-phytyl-1,4-benzoquinone methyltransferase (plastoquinol and tocopherol biosynthesis). These results indicated that the main carotenoid biosynthesis pathway, which proceeds through IPP and GGPP, was active and that the synthesis of other metabolites was partially blocked by the overexpression of CsMADS6.
Plant hormones, as a class of small molecules present in low abundance, play important roles at various points in the life cycles of plants. RNA-seq data showed that the transcript levels of genes involved in hormone biosynthesis were changed significantly. For example, the transcript levels of 1-aminocyclopropane-1-carboxylate oxidase1 and 1-aminocyclopropane-1-carboxylate synthase2/4, which encode enzymes for ethylene biosynthesis, and CYP85A (dwarf, brassinosteroid-6-oxidase), which encodes an enzyme involved in brassinosteroid biosynthesis, were expressed at much higher levels in transgenic sepals than in the controls.
Many DEGs that were annotated as TFs and regulators were differentially expressed in the M6SvsWTS comparison. The most differentially expressed regulatory genes belonged to the AP2/ERF, MADS, MYB, bHLH, and WRKY families (Supplemental Fig. S7). The expression levels of many fruit ripening-related TFs, such as RIN, FUL, and NOR (nonripening), were up-regulated significantly in the transgenic sepals. We also observed the down-regulation of other fruit ripening-related TFs, such as HD-Zip homeobox and GLK1. The expression of TAGL1, a CsMADS6 homolog in tomato, did not vary significantly.
DISCUSSION
Carotenoids are important secondary metabolites in plants. LCYb1 is known to play an important role in plant carotenoid metabolism, but the transcriptional regulation of LCYb1 is not well understood. Citrus fruit is rich in carotenoids. Thus, understanding the mechanisms that regulate carotenoid metabolism is of great significance to the citrus industry. In this study, we identified a citrus MADS TF, CsMADS6, with potential roles in regulating the expression of LCYb1 and the accumulation of carotenoids. CsMADS6 belongs to the AGAMOUS-like subfamily and is homologous to the TAGL1 protein from tomato (Fig. 1). Previous studies have reported that the carotenoid profiles and the transcript levels of biosynthetic genes are influenced significantly by the overexpression or suppression of tomato TAGL1 (Itkin et al., 2009; Vrebalov et al., 2009). In addition, we found that CsMADS6 was expressed strongly in a ripening-specific manner (i.e. associated with the accumulation of carotenoids in citrus fruit; Fig. 2; Liu et al., 2007). These data are consistent with CsMADS6 contributing to carotenoid metabolism. We subsequently obtained direct evidence to support this idea.
CsMADS6 Is a Direct and Positive Regulator of LCYb1
Using a variety of approaches (i.e. Y1H, EMSA, and dual-luciferase assay), we first demonstrated that CsMADS6 binds directly to the promoter of LCYb1 and activates the promoter activity of LCYb1 (Fig. 4). In addition, we found that the transcript abundance of LCYb1 was significantly higher in CsMADS6-overexpressing citrus calli (Fig. 5D) and CsMADS6-overexpressing tomato pericarps (Fig. 9A) than in the controls, which provided more evidence that CsMADS6 induced the expression of LCYb1. Previous studies have reported that the expression of LCYb genes is higher in tomato TAGL1-RNA interference lines, which is consistent with TAGL1 negatively regulating the expression of LCYb (Vrebalov et al., 2009). However, in this study, the transcript levels of LCYb genes were higher in CsMADS6-overexpressing tomato pericarps than in the control (Fig. 9A), which is consistent with CsMADS6 positively regulating the expression of LCYb in tomato pericarps. These findings indicated that CsMADS6 and its homolog (TAGL1) both regulated the expression of LCYb in the pericarp of tomato, but the nature of their regulatory activity was distinct. Citrus fruits mainly accumulate xanthophylls, such as violaxanthin and β-cryptoxanthin, but rarely accumulate lycopene (Kato et al., 2004; Fanciullino et al., 2006). During the development of citrus fruit, the transcript levels of LCYb genes increase gradually, and the expression patterns of LCYb and CsMADS6 were similar (Kato et al., 2004; Liu et al., 2007; Lu et al., 2016b). In contrast, during the development of tomato fruit, the expression of LCYb genes was down-regulated gradually, and the expression patterns of LCYb and TAGL1 were inversely associated. The down-regulation of LCYb expression is followed by the accumulation of large quantities of lycopene in fruit (Pecker et al., 1996; Ronen et al., 1999, 2000). Therefore, it is reasonable to suggest that LCYb expression is positively regulated by CsMADS6 and negatively regulated by TAGL1, showing the species-specific regulation of LCYb expression by AGAMOUS-like MADS TFs.
CsMADS6 Promotes Carotenoid Metabolism by Directly Regulating the Expression of LCYb1 and Other Carotenogenic Genes, Including PSY, PDS, and CCD1
This study found that the overexpression of CsMADS6 resulted in increased β-branch carotenoid content in both transgenic citrus calli (Fig. 5C) and tomato pericarps (Fig. 7C). It is difficult to suppress fruit-specific MADS genes in citrus, which are perennial woody trees. However, previous studies have reported that silencing its homolog in tomato leads to significantly lower amounts of carotenoids in fruit, and these changes correlate with the reduction in the transcript levels of PSY and other biosynthetic genes (Itkin et al., 2009; Vrebalov et al., 2009). Collectively, these findings indicate that CsMADS6 positively modulates carotenoid metabolism. This study discovered that many carotenogenic genes other than LCYb1, such as PSY, were affected by the overexpression of CsMADS6 (Figs. 5D and 9A). PSY is generally accepted as a key determinant of total carotenoids in a number of fruits, such as tomato (Fraser et al., 1994; Bramley, 2002) and citrus fruits (Kato et al., 2004; Tao et al., 2007). In this study, we found that the levels of total carotenoids were enhanced consistently when the transcript levels of PSY were enhanced in the CsMADS6-overexpressing lines. The high levels of LCYb1, LCYb2, and HYD, together with the reduced levels of LCYe, directed lycopene into the β-branch of carotenoid synthesis, resulting in elevated levels of violaxanthin in CsMADS6-overexpressing citrus calli (Fig. 5C), which resembles the carotenogenesis that occurs in the ripening fruit of sweet orange (Kato et al., 2004; Liu et al., 2007). In transgenic tomato, the altered expression levels of carotenoid biosynthetic genes similarly increased the levels of the β-branch carotenoids in CsMADS6-overexpressing lines (Fig. 7C). In contrast to the greatly increased transcript levels from the biosynthetic genes, especially PSY and PDS, carotenoid profiles changed little in transgenic calli. Lätari et al. (2015) determined that the unchanged carotenoid levels in the leaves of Arabidopsis AtPSY-overexpressing lines were due to CCD4 cleaving specific xanthophyll molecules to yield high levels of C13 apocarotenoid glycosides. Thus, the discordance produced by this study may be partially explained by the increased transcript levels of CCD1, which catalyzes the enzymatic degradation of carotenoids to yield volatile compounds such as β-cyclocitral or other apocarotenoids (Simkin et al., 2004). It is difficult to detect these derivatives because of their low content in citrus calli.
Overexpressing CsMADS6 in tomato sepals dramatically altered the carotenoid profiles and the expression of many carotenoid-associated genes (Figs. 7D and 9B). Similar phenomena have been reported in previous studies of homologous genes from tomato and peach, which is consistent with the conservation of the transcriptional control of fruit ripening by the AGAMOUS-like MADS regulators in climacteric and nonclimacteric species (Itkin et al., 2009; Tadiello et al., 2009; Vrebalov et al., 2009). The extent of the changes was relatively greater in the transgenic sepals than in the transgenic pericarps. We also discovered that overexpression of CsMADS6 had different effects on gene expression in transgenic pericarps and sepals (Fig. 9). For example, tomato LCYb1 was significantly up-regulated in transgenic pericarps but down-regulated in transgenic sepals. These findings are consistent with the tissue type influencing the transcriptional regulation by CsMADS6. Jeong et al. (2010) reported that the number of genes up-regulated by OsNAC10 in rice (Oryza sativa) is different in roots and leaves and that the expression of only four genes is up-regulated in both organs. Alternatively, Bovy et al. (2002) reported that the LC/C1 TF effect on flavonoid gene expression is much more modest in leaves than in fruit because the flavonoid pathway is already active in wild-type leaves. Similarly, the moderate changes in transgenic tomato pericarps may be due to the high activity of the carotenoid biosynthetic pathway in tomato pericarps. We cannot rule out the possibility that this expression in tomato sepals and the significantly different expression of other ripening-related TFs, which were revealed by RNA-seq data, are coordinately regulated (Fig. 10).
Further investigation demonstrated that CsMADS6 could bind directly to the promoters and up-regulate the expression of PSY, PDS, and CCD1 (Fig. 6). In the dual-luciferase assays, we found that CsMADS6 induced higher levels of activation of the CCD1 promoter than those of the PSY and PDS promoters (Fig. 6D). Espley et al. (2009) demonstrated that, in the presence of the MYB10 effector, the expression of the LUC reporter increases when the number of cis-acting elements in the promoter increases. The numbers of the predicted CArG elements in the promoter of CCD1 (five) were more than those of PSY (two) and PDS (three; Supplemental Table S2), possibly suggesting a positive correlation between the number of CArG elements and the activation of the promoter by CsMADS6. Our results of the dual-luciferase assays displayed the ability of CsMADS6 to activate the two promoter fragments of LCYb1 (LP1 and LP2). However, a deletion from LP1 to LP2 decreased the numbers of CArG elements, leading to high levels of activation by CsMADS6. These data imply that the sequence context located from LP1 to LP2 may affect the binding affinity or activation of CsMADS6 to the promoter of LCYb1. The increased expression of CRTISO, LCYb2, and HYD in transgenic lines also may be regulated by CsMADS6 through the possible CArG elements discovered in their promoters (Supplemental Table S2). Hu et al. (2016) identified an HD-ZIP TF, SlHZ24, that modulates ascorbate levels by targeting the genes encoding three members of the d-Man/l-Gal biosynthetic pathway. Chromatin immunoprecipitation sequencing analyses indicate that the tomato FUL homologs generally regulate carotenoid metabolism and that RIN specially regulates the accumulation of lycopene (Fujisawa et al., 2013, 2014). This study clearly shows that, similar to the FUL homologs, CsMADS6 affects carotenoid metabolism by directly regulating the expression of multiple genes that contribute to carotenoid biosynthesis.
In this study, we found that the expression of HY5, RAP2.2, and PIF1 changed significantly in transgenic citrus calli (Supplemental Fig. S1). We propose that CsMADS6 may directly target the genes encoding these three TFs because of the presence of CArG elements in their promoters (Supplemental Table S2). Previous studies demonstrate that PIF1 binds directly to PSY and down-regulates its expression. In contrast, HY5 acts antagonistically to activate PSY expression (Toledo-Ortiz et al., 2010, 2014). In the Arabidopsis root-derived calli, the decrease in AtRAP2.2 transcript levels in a T-DNA insertion line leads to the down-regulated expression of PSY and PDS, which have the AtRAP2.2-binding motif in their promoters (Welsch et al., 2007). Based on these data, we suggest that the high expression of PSY and PDS, which have the binding sites for HY5, RAP2.2, and PIF1 in their promoters (Supplemental Table S2), is enhanced synergistically by the elevated expression of both HY5 and RAP2.2 and the repression of PIF1 in transgenic citrus calli. Moreover, previous studies reported that carotenoids and their derivatives, such as β-cyclocitral and dihydroactinidiolide, act as retrograde signals to regulate the expression of genes in the nucleus (Ramel et al., 2012; Shumbe et al., 2014). The expression of Daucus carota DcLCYb1 in tobacco (Nicotiana tabacum) induces a positive feedback affecting the expression of key carotenogenic genes, NtPSY1, NtPSY2, and NtLCYb (Moreno et al., 2016). These data permit us to propose that the significantly increased transcript levels of PSY and PDS that are a consequence of CsMADS6 expression may result from positive feedback regulation, mediated by a molecule produced by the increased expression of CCD1, which cleaves β-carotene and violaxanthin (Fig. 5, C and D).
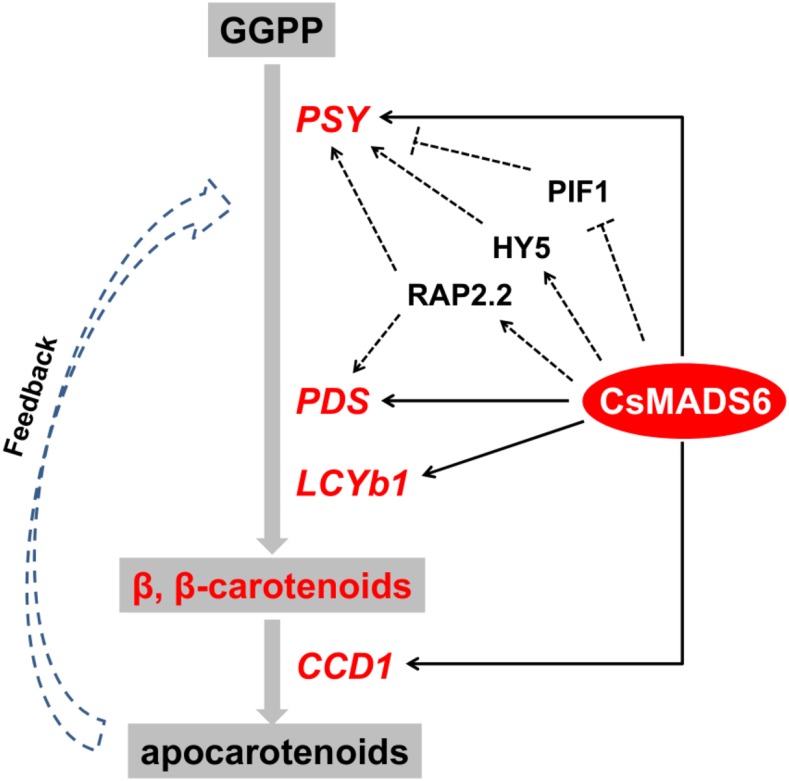
Based on the results presented here and elsewhere, we propose a model to elucidate the transcriptional regulation of carotenoid metabolism by CsMADS6 (Fig. 11). CsMADS6 coordinately and positively modulates carotenoid metabolism by directly regulating the expression of LCYb1, PSY, PDS, and CCD1. The up-regulated expression of PSY, PDS, and LCYb1, together with the altered expression of other carotenogenic genes, induces carotenoid biosynthesis and directs flux into the β-branch, consequently forming more β,β-carotenoids in the transgenic lines. The up-regulated expression of CCD1 balances carotenoid content and composition and also permits the production of apocarotenoids, such as β-cyclocitral, which may act as retrograde signal(s) that regulate the expression of carotenogenic genes. Moreover, CsMADS6 probably induces the notably high expression of PSY and PDS by an indirect mechanism that involves regulating the expression of HY5, RAP2.2, and PIF1. The feedback regulation by apocarotenoids and the indirect regulation by other TFs (indicated with dashed lines in Fig. 11) remain to be determined. This model enriches our understanding of the complex transcriptional regulation of carotenoid metabolism in plants.
Figure 11.
Proposed model for the transcriptional regulation of carotenoid metabolism by CsMADS6. Thick gray arrows indicate the simplified reaction flows in the pathway. The biosynthetic genes encoding enzymes that catalyze the reactions are shown at the sides of the arrows. CsMADS6 coordinately and positively modulates carotenoid metabolism by directly regulating the expression of LCYb1, PSY, PDS, and CCD1. The up-regulated expression of PSY, PDS, and LCYb1, together with the altered expression of other carotenogenic genes, induces carotenoid biosynthesis and directs flux into the β-branch, consequently forming more β,β-carotenoids in the transgenic lines. The up-regulated expression of CCD1 balances carotenoid content and composition and also permits the production of apocarotenoids, such as β-cyclocitral, which may act as retrograde signals that regulate the expression of carotenogenic genes. Moreover, CsMADS6 probably induces the notably high expression of PSY and PDS by an indirect mechanism that involves regulating the expression of HY5, RAP2.2, and PIF1. The feedback regulation by apocarotenoids and the indirect regulation by other TFs (indicated with dashed lines) remain to be determined. This model enriches our understanding of the complex transcriptional regulation of carotenoid metabolism in plants.
CsMADS6 Reprograms Transcriptional Networks to Direct Metabolic Flux into the Carotenoid Pathway
Global transcriptome analysis of transgenic tomato sepals identified many genes whose expression was affected significantly by CsMADS6 overexpression (Supplemental Table S3). Consistent with the observed phenotypic changes, genes related to chlorophyll metabolism and photosynthesis were down-regulated significantly, while the expression of genes associated with carotenoid biosynthesis was increased dramatically. The transcript levels of key genes, such as NCED and D27, involved in the enzymatic degradation of carotenoids were repressed significantly in the sepals of transgenic tomato. Sun et al. (2012) reported that suppression of SlNCED1 produces deep-red fruit with increased accumulation of lycopene and β-carotene. We propose that the notably increased lycopene content in transgenic sepals could not only be a result of enhanced carbon flow to carotenoid production but also of the inhibition of carotenoid degradation. Furthermore, this study found that the plastids in transgenic sepals developed into chromoplast structures (Fig. 8), which are responsible for the attractive colors of fruits and flowers (Vishnevetsky et al., 1999; Li and Yuan, 2013). Considering these results, the CsMADS6 protein is involved in three processes, promoting biosynthesis, repressing degradation, and stabilizing storage, to facilitate specific carotenoid (lycopene) accumulation in transgenic tomato sepals.
Large-scale transcriptome analysis identified many DEGs involved in other biological processes and metabolic pathways (Fig. 10; Supplemental Table S4). For example, two key genes, PK and PFK, which are involved in primary metabolism, were up-regulated significantly, resulting in the production of pyruvate, an important intermediate linking primary and secondary metabolism. The expression of genes encoding enzymes that catabolize pyruvate by anaerobic respiration or through the tricarboxylic acid cycle was repressed. In contrast, the expression of genes that contribute to the MVA and MEP pathways, which supply precursors for the biosynthesis of secondary metabolites, was induced significantly. The expression of genes associated with the pathways of secondary metabolism other than the carotenoid biosynthetic pathway was repressed significantly. These findings indicate that CsMADS6 can regulate the expression of carotenoid biosynthetic genes and reprogram carbon flux toward carotenoid synthesis. Similarly, Zhang et al. (2015) showed that the AtMYB12 TF not only increased flavonoid biosynthesis but also increased primary metabolism to fuel Phe biosynthesis, thus enabling effective production of phenylpropanoids in transgenic tomato fruit. Previous studies reported that phytohormones, such as ethylene (Klee and Giovannoni, 2011) and brassinosteroids (Lanahan et al., 1994; Liu et al., 2014), and TFs could act cooperatively and independently to promote carotenoid biosynthesis (Giovannoni, 2007; Gapper et al., 2013). This study showed that the expression of key genes involved in ethylene and brassinosteroid biosynthesis increased significantly. Moreover, the expression of many fruit ripening-related genes that encode TFs, such as RIN, NOR, FUL1, and FUL2, changed in transgenic sepals. These findings indicate that the increased levels of ethylene and brassinosteroid biosynthetic genes, as well as those of the fruit ripening-related TFs, may assist in the accumulation of carotenoids. A previous study reported that the endogenous activity of TAGL1 might be interfered with by the ectopic expression of peach PpPLENA (a TAGL1 homolog; Tadiello et al., 2009). Our study found that TAGL1 expression was not altered significantly in transgenic tomato, implying that the transcriptional changes in transgenic sepals are regulated by CsMADS6 and not its homolog from tomato. However, the roles of other TFs that were affected by CsMADS6 overexpression certainly cannot be excluded. Later, we will conduct transcriptome analysis of transgenic citrus calli to avoid the possibly indirect effects caused by these ripening regulators and to deeply understand the transcriptional regulatory functions of citrus CsMADS6.
Based on the above analysis, we provide a schematic of a global view of transcriptional regulation by CsMADS6 (Fig. 10B). This view shows that CsMADS6 coordinately regulates a series of pathways, including primary pathways, secondary pathways, and phytohormone and TF levels, to synergistically promote the accumulation of large amounts of a specific carotenoid (lycopene) in transgenic sepals. The multiple regulatory functions of CsMADS6 indicate that it plays important roles in the reprogramming of transcriptional networks to direct metabolic flux into the carotenoid pathway.
CONCLUSION
In conclusion, we identified a citrus MADS TF named CsMADS6 that regulates carotenoid metabolism by directly regulating the expression of carotenogenic genes. Moreover, CsMADS6 coordinates many other metabolic pathways to synergistically promote the biosynthesis and accumulation of carotenoids. The manipulation of CsMADS6 expression, therefore, may be useful for manipulating the carotenoid content in citrus and other fruits. Thus, this study describes a new MADS regulon that enables our understanding of the physiological relevance of MADS proteins in carotenoid metabolism. In the future, we will combine transcriptome analysis with chromatin immunoprecipitation assays to thoroughly identify the direct targets of CsMADS6 to better understand carotenoid metabolism in plants. Moreover, we will attempt to analyze sequence polymorphisms and the expression of CsMADS6 among different citrus varieties that exhibit different fruit colors and use CRISPR/Cas9 to knock down CsMADS6 in citrus to learn more about its role in the regulation of carotenoid metabolism in citrus fruit.
MATERIALS AND METHODS
Plant Materials
The citrus materials used in this study were from the ‘Hong Anliu’ sweet orange (Citrus sinensis). Roots, stems, and leaves were sampled from six-leaf-stage plantlets. Fruits at six developmental stages (60, 90, 120, 150, 180, and 220 DAF) were collected from mature trees growing in the orchard of the National Citrus Breeding Center at Huazhong Agricultural University. All samples were frozen immediately in liquid nitrogen and stored at −80°C until they were used.
Gene Isolation and Sequence Analysis
The full-length coding sequence (CDS) of CsMADS6 was amplified using PCR with the primers listed in Supplemental Table S5. The conserved domains were analyzed using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd) and the ExPASy Prosite Database (http://prosite.expasy.org/). A set of associated MADS protein sequences was downloaded from GenBank or other genome databases. Multiple sequence alignments were prepared using ClustalX and visualized in GeneDoc. A phylogenetic tree was constructed using MEGA software with the neighbor-joining method. The reliability of the nodes in the tree was evaluated by bootstrapping with 1,000 replicates.
RNA Extraction and RT-qPCR
Total RNA was extracted from all samples using the TRIzol Reagent (Life Technologies). First-strand cDNA was synthesized using the HiScript II First Strand cDNA Synthesis Kit (+gDNA wiper; Vazyme). RT-qPCR was performed with the Roche LightCycler 480 system using the 2× LightCycler 480 SYBR Green master mix (Roche) and a three-step program: preincubation at 95°C for 10 min; 40 cycles of amplification at 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s; followed by a melting curve at 95°C for 5 s, 65°C for 1 min, and then ramping at 0.11°C s−1 to 97°C with continuous fluorescence measurement. Gene-specific primers used for RT-qPCR were designed using PrimerExpress software. Actin was chosen as the endogenous control. Fluorescence was measured at each extension step. Each run contained a negative control (water in place of cDNA), and each reaction was performed in triplicate. The reaction specificity was confirmed by the negative control and a melting temperature calling analysis. The data were analyzed using LightCycler 480 software release 1.5.0 (Roche).
Protein Extraction and Immunoblotting
The CsMADS6 protein was detected in extracts prepared from cv Hong Anliu fruits at six developmental stages (60, 90, 120, 150, 180, and 220 DAF). Anti-CsMADS6 polyclonal antibody was commissioned from Frdbio. Protein extraction and immunoblotting were performed using the methods described by Pan et al. (2009) and Cao et al. (2012) with minor modifications.
Subcellular Localization
The CDS from CsMADS6 without the stop codon was amplified using PCR and cloned into the PM999 vector in frame with the GFP gene. Citrus leaf protoplasts were cotransformed with the plasmid encoding CsMADS6-GFP and a plasmid encoding a nuclear marker, OsGhd7-CFP, using a polyethylene glycol-based method. Florescence signals were observed with a confocal laser scanning microscope (TCS SP2; Leica) 20 h after transformation.
Y1H Analysis
Y1H screening was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clotech). The promoter fragments from citrus LCYb1 and LCYb2 were cloned into the pAbAi vector to produce the bait constructs pAbAi-LCYb1 and pAbAi-LCYb2, respectively. The bait plasmids were then linearized and integrated into the Y1HGold yeast (Saccharomyces cerevisiae) genome to create bait strains. A citrus fruit cDNA library was constructed using SMART cDNA synthesis technology. The cDNA library was then transformed into the bait yeast cells and selected on SD plates lacking Leu with or without AbA. The prey fragments from the positive colonies were identified by DNA sequencing (Augct).
For the Y1H assay, different promoter fragments or three copies of the CArG element were cloned separately into the pAbAi vector to create bait constructs. The CDS of CsMADS6 was fused to the GAL4 AD in the pGADT7 vector to generate a prey construct, AD-CsMADS6. The prey vector and the empty vector (AD), serving as the negative control, were then transformed separately into yeast cells containing bait constructs. The transformed yeast cells were diluted with a 10× dilution series and dotted on the SD plates lacking Leu with or without antibiotic. The cells that grew on both media contained prey proteins and the bait sequences that interacted.
EMSA
The CDS of CsMADS6 without the stop codon was cloned into a double-tagged expression vector to generate the recombinant vector MBP-CsMADS6-His. Escherichia coli BL21 (DE3) was transformed with this construct for the expression of recombinant protein. The recombinant protein was expressed and purified using Ni-NTA agarose columns. The promoter fragments containing the CArG elements or three copies of the potential CArG elements were synthesized and labeled with FAM or Cy5 luciferase. Complementary oligonucleotides were annealed and used as EMSA probes. Unlabeled probes with the same or mutated oligonucleotides were used as cold competitors. EMSA binding reactions were performed according to the manufacturer’s instructions (LightShift EMSA Kit; Thermo Scientific). Cold competitor DNA was added to at least a 20-fold excess relative to the labeled probe. The binding complexes were then loaded onto a prerunning native 6% (w/v) polyacrylamide gel, and electrophoresis was performed at a low temperature using 0.5× Tris-borate-EDTA buffer at 100 V for 1 h. Gel images were captured using the Amersham Imager 600 (GE Healthcare).
Transient Expression Assay
For the transcriptional activity assay, the reporter vector, a derivative of pGreenII 0800-LUC, containing five copies of the GAL4 AD upstream of the minimal CaMV35S promoter and the LUC gene, was used. For an internal control, the expression of the REN gene was driven by the CaMV35S promoter in a reporter vector. The CDS of CsMADS6 was inserted upstream of the GAL4 DNA-binding domain in the pBD vector to generate an effector vector named pBD-CsMADS6. The empty vector was used as the negative control (pBD), while the vector containing the VP16 activation domain was used as the positive control (pBD-VP16).
For the DNA-promoter interaction assay, promoter sequences were amplified from genomic DNA using PCR and inserted upstream of the LUC CDS in pGreen 0800-LUC to yield the promoter-LUC reporter vectors. The CDS of CsMADS6 was cloned into the pK2GW7 destination vector using the Gateway Cloning System (Invitrogen) to create an overexpression construct named pK2-CsMADS6. The empty vector pK2GW7 was used as the negative control (pK2).
All resulting constructs were introduced into the Agrobacterium tumefaciens strain EHA105 using the freeze-thaw method or strain GV3101 by electroporation. Transient expression in Nicotiana benthamiana was performed according to the method described by Hellens et al. (2005). Luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega) with an Infinite200 Pro microplate reader (Tecan). Expression was expressed as the ratio of LUC to REN activity and normalized to the negative control.
Transformation of Citrus Calli and Tomato Plants
An A. tumefaciens strain containing the overexpression vector pK2 or pK2-CsMADS6 was used in the plant transformation experiment. The transformation of citrus calli and growth conditions were as described previously (Lu et al., 2016a). The transformation of tomato (Solanum lycopersicum ‘Ailsa Craig’) was performed as described by Sun et al. (2006) with minor modifications by Bee Lynn Chew and Yu Pan (University of Nottingham). After their roots were established, the transgenic plants were transferred to soil and grown in a glasshouse alongside untransformed control tomato plants. Genomic PCR using specific primers for both the antibiotic resistance gene and the target gene was performed to verify positive transformations. Expression levels were examined by RT-PCR and immunoblotting. Insertions at single loci were identified using quantitative real-time PCR. Transgene copy numbers were estimated by reference to the endogenous single-copy early light-inducible protein gene. Overexpression of CsMADS6 affected reproductive development and reduced both fruit set and seed numbers per fruit. Therefore, the subsequent physiological and biochemical determinations were carried out in the T1 generation. Tomato fruit samples were collected from the T1 progeny of three independent transgenic lines that contained single-copy transgene insertions and had high expression of CsMADS6 and were analyzed further.
Quantitation of Carotenoid and Chlorophyll Levels
Lyophilized samples were used for physiological measurements. Carotenoid extraction and determination were performed as described previously (Cao et al., 2012). Carotenoids were analyzed using reverse-phase HPLC, which was performed in a Waters liquid chromatography system equipped with a model 1525 solvent delivery system, a model 2998 photodiode array detection system, a model 2707 plus autosampler, and an Empower Chromatography Manager. The carotenoids were eluted from a C30 carotenoid column (250 × 4.6 mm; YMC). Carotenoids were identified by their characteristic absorption spectra and retention times based on the literature and standards purchased from CaroteNature. Carotenoids were quantified using calibration curves that were prepared for violaxanthin, lutein, β-carotene, and lycopene. The levels of α-carotene in citrus calli were calculated with the peak area: [peak area × volume (μL)]/[weight (g) × 106]. The peak areas of lycopene were quantified at the wavelength of 471 nm, while those of other carotenoids were quantified at 450 nm. The UV spectra for all of the carotenoids identified in this study are provided in Supplemental Figure S8.
Chlorophyll (Chl) extraction and determination were performed as described by Wellburn (1994). The A663 (A663) and A645 (A645) were determined using the Infinite200 Pro microplate reader (Tecan). We used the following formulae: total Chl (mg g−1) = (8.04A663 + 20.2A645) × volume (L)/dry weight (g); Chl a (mg g−1) = (12.72A663 – 2.69A645) × volume (L)/dry weight (g); and Chl b (mg g−1) = (22.89A645 – 4.68A663) × volume (L)/dry weight (g).
TEM
TEM was performed as described previously (Cao et al., 2012, 2015). Fruit sepals and pericarps were collected from wild-type and transgenic tomato plants at the mature fruit stage. All samples were hand dissected into small squares. Excised tissues were then immersed immediately in fixative solutions and sent to the electron microscopy platform of Huazhong Agricultural University for subsequent manipulations and procedures. TEM was performed with a HITA-CHI H-7650 transmission electron microscope at 80 kV and a Gatan 832 CCD camera.
RNA Library Construction and Sequencing
Three groups were sequenced (wild-type sepals [WTS], wild-type pericarps [WTR], and CsMADS6-overexpressing sepals [M6S]). For each group, three independent biological replicates were subjected to RNA-seq. All samples were collected at the Br+5 stage of fruit development. RNA library construction and sequencing were conducted by Novogene Bioinformatics Technology using Illumina HiSeq. HTSeq version 0.6.1 software in union mode was used to count the read numbers mapped to each gene (Anders et al., 2015). The gene expression levels were calculated using the commonly used FPKM (expected number of fragments per kilobase of transcript sequence per million bp sequenced) method (Trapnell et al., 2010). The RNA-seq quality and read numbers are summarized in Supplemental Table S6. The criterion of an adjusted P < 0.05 was used to search DEGs, which are listed in Supplemental Table S3. GO and KEGG enrichment analyses were implemented using the GOseq R package and KOBAS software, respectively (Mao et al., 2005; Young et al., 2010). HemI 1.0 software was used for the hierarchical clustering analysis and heat map illustration (Deng et al., 2014). The CArG element in the promoter was searched using the Softberry Nsite-PL program (http://linux1.softberry.com/). Raw data were deposited in the NCBI database under accession number PRJNA432771.
Statistical Analysis
The data are presented as means ± sd of at least three independent experiments. Statistical analyses were conducted using the one-way ANOVA test with the Microsoft Excel program (Microsoft Office 2010). Statistically significant differences are indicated with either single asterisks or lowercase letters (P < 0.05) and double asterisks or uppercase letters (P < 0.01).
Accession Numbers
Sequence data from this article can be found in the NCBI database or in the genome databases of citrus (http://citrus.hzau.edu.cn/orange/) or Arabidopsis (http://www.arabidopsis.org/index.jsp) under the following accession numbers: Citrus sinensis CsMADS6 (MG594037) and CsMADS1 (MG594038); Arabidopsis thaliana AtAG (AT4G18960), AtSHP1 (AT3G58780), AtSHP2 (AT2G42830), and AtFUL (AT5G60910); Antirrhinum majus AmFAR (CAB42988) and AmPLENA (BAI68391); Solanum lycopersicum SlTAG1 (AAA34197), SlTAGL1 (NP_001300859), and SlRIN (AAM15775); Nicotiana tabacum NtAG1 (AAA17033) and NtSHP (AFK13160); Vitis vinifera VvMADS1 (AAK58564); Prunus persica PpPLENA (ACL31234); and Fragaria × ananassa FaSHP (AGU92563). The accession numbers of the genes in the RT-qPCR analysis are provided in Supplemental Table S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative expression levels of HY5, RAP2.2, and PIF1 in transgenic citrus calli.
Supplemental Figure S2. Chlorophyll content in transgenic tomato sepals at the Br+5 stage of fruit development.
Supplemental Figure S3.Volcano plots showing the whole distribution of DEGs in the M6SvsWTS and M6SvsWTR comparisons.
Supplemental Figure S4. Correlation analysis of DEGs obtained from RNA-seq and RT-qPCR assays.
Supplemental Figure S5. Hierarchical clustering analysis of transcript levels obtained by RNA-seq in WTS, M6S, and WTR.
Supplemental Figure S6. GO terms and KEGG classifications of DEGs in the M6SvsWTS and M6SvsWTR comparisons.
Supplemental Figure S7. TF families of DEGs in the M6SvsWTS and M6SvsWTR comparisons.
Supplemental Figure S8. HPLC analysis of carotenoids in CsMADS6-overexpressing lines.
Supplemental Table S1. A list of interacting factors identified by Y1H screening.
Supplemental Table S2. Related cis-elements predicted in the promoters of several carotenoid-associated genes from citrus.
Supplemental Table S3.DEGs from the M6SvsWTS and M6SvsWTR comparisons.
Supplemental Table S4. Selected carotenoid-associated DEGs from the M6SvsWTS comparison.
Supplemental Table S5. Primers and probes used in this study.
Supplemental Table S6. Summary of read numbers based on the RNA-seq data.
Acknowledgments
We greatly appreciate the time and effort of Robert M. Larkin (Huazhong Agricultural University) in editing the English language of this article. We thank Jihong Liu (Huazhong Agricultural University), Yujin Hao (Shandong Agricultural University), Qiang Zhao (Chinese Academy of Agricultural Sciences), and Xiongjie Zheng (Huazhong Agricultural University) and Jinlong Wu (Huazhong Agricultural University) for their comments and suggestions on this article. We thank Juan Xu (Huazhong Agricultural University) and Meiyan Shi (Huazhong Agricultural University) for providing standard curves for carotenoids; Andrew C. Allan (New Zealand Institute for Plant and Food Research) for providing vectors for the dual-luciferase assay; Wangjin Lu (South China Agricultural University) and Jianfei Kuang (South China Agricultural University) for providing vectors for the transcriptional activation assay; and Lizhong Xiong (Huazhong Agricultural University) for providing vectors for the subcellular localization experiments. We also thank Hongyan Zhang (Huazhong Agricultural University) for technical assistance; Junhong Zhang (Huazhong Agricultural University) and Bo Ouyang (Huazhong Agricultural University) for providing tomato seeds and plantations; Huiyang Yu (Huazhong Agricultural University) and Guo Ai (Huazhong Agricultural University) for guidance in tomato management; and Huiyang Yu (Huazhong Agricultural University) and Yue Huang (Huazhong Agricultural University) for assistance in the bioinformatics analysis of the RNA-seq data.
Footnotes
This research was financially supported by the National Natural Science Foundation of China (nos. 31330066, 31521092, and 31630065), the China Agriculture Research System (CARS-27), and the 111 project (B13034).
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Alquézar B, Zacarías L, Rodrigo MJ (2009) Molecular and functional characterization of a novel chromoplast-specific lycopene β-cyclase from Citrus and its relation to lycopene accumulation. J Exp Bot 60: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA (1995) Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto PdB, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al. (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley PM. (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53: 2107–2113 [DOI] [PubMed] [Google Scholar]
- Burns J, Fraser PD, Bramley PM (2003) Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry 62: 939–947 [DOI] [PubMed] [Google Scholar]
- Cao H, Wang J, Dong X, Han Y, Ma Q, Ding Y, Zhao F, Zhang J, Chen H, Xu Q, et al. (2015) Carotenoid accumulation affects redox status, starch metabolism, and flavonoid/anthocyanin accumulation in citrus. BMC Plant Biol 15: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Zhang J, Xu J, Ye J, Yun Z, Xu Q, Xu J, Deng X (2012) Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J Exp Bot 63: 4403–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Cunningham FX Jr, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E (1996) Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS ONE 9: e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mascio P, Kaiser S, Sies H (1989) Lycopene as the most efficient biological carotenoid singlet oxygen quencher. Arch Biochem Biophys 274: 532–538 [DOI] [PubMed] [Google Scholar]
- Dong T, Hu Z, Deng L, Wang Y, Zhu M, Zhang J, Chen G (2013) A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol 163: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge R, McGarvey DJ, Truscott TG (1997) The carotenoids as anti-oxidants: a review. J Photochem Photobiol B 41: 189–200 [DOI] [PubMed] [Google Scholar]
- Endo T, Fujii H, Sugiyama A, Nakano M, Nakajima N, Ikoma Y, Omura M, Shimada T (2016) Overexpression of a citrus basic helix-loop-helix transcription factor (CubHLH1), which is homologous to Arabidopsis activation-tagged bri1 suppressor 1 interacting factor genes, modulates carotenoid metabolism in transgenic tomato. Plant Sci 243: 35–48 [DOI] [PubMed] [Google Scholar]
- Endo T, Shimada T, Fujii H, Omura M (2006) Cloning and characterization of 5 MADS-box cDNAs isolated from citrus fruit tissue. Sci Hortic (Amsterdam) 109: 315–321 [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciullino AL, Dhuique-Mayer C, Luro F, Casanova J, Morillon R, Ollitrault P (2006) Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. J Agric Food Chem 54: 4397–4406 [DOI] [PubMed] [Google Scholar]
- Fiedor J, Burda K (2014) Potential role of carotenoids as antioxidants in human health and disease. Nutrients 6: 466–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CC, Han YC, Fan ZQ, Chen JY, Chen WX, Lu WJ, Kuang JF (2016) The papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J Agric Food Chem 64: 5454–5463 [DOI] [PubMed] [Google Scholar]
- Fu CC, Han YC, Kuang JF, Chen JY, Lu WJ (2017) Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4, CpLCY-e, and CpCHY-b during fruit ripening. Plant Cell Physiol 58: 2155–2165 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y (2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y (2014) Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ (2013) Molecular and genetic regulation of fruit ripening. Plant Mol Biol 82: 575–591 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Han Y, Wu M, Cao L, Yuan W, Dong M, Wang X, Chen W, Shang F (2016) Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol Biol 91: 485–496 [DOI] [PubMed] [Google Scholar]
- Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M (2015) Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genet 11: e1005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt NE, Zigmantas D, Valkunas L, Li XP, Niyogi KK, Fleming GR (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Hu T, Ye J, Tao P, Li H, Zhang J, Zhang Y, Ye Z (2016) The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J 85: 16–29 [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134: 824–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1994) The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lätari K, Wüst F, Hübner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R (2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H (2013) Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys 539: 102–109 [DOI] [PubMed] [Google Scholar]
- Liu L, Jia C, Zhang M, Chen D, Chen S, Guo R, Guo D, Wang Q (2014) Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol J 12: 105–115 [DOI] [PubMed] [Google Scholar]
- Liu L, Shao Z, Zhang M, Wang Q (2015) Regulation of carotenoid metabolism in tomato. Mol Plant 8: 28–39 [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu Y, Zhao X, Deng X, Guo L, Gu J (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J Exp Bot 58: 4161–4171 [DOI] [PubMed] [Google Scholar]
- Llorente B, D’Andrea L, Ruiz-Sola MA, Botterweg E, Pulido P, Andilla J, Loza-Alvarez P, Rodriguez-Concepcion M (2016) Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J 85: 107–119 [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang Y, Zheng X, Zhu K, Xu Q, Deng X (2016a) Isolation and functional characterization of a lycopene β-cyclase gene promoter from citrus. Front Plant Sci 7: 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SW, Zhang Y, Zheng XJ, Zhu KJ, Xu Q, Deng XX (2016b) Molecular characterization, critical amino acid identification, and promoter analysis of a lycopene β-cyclase gene from citrus. Tree Genet Genomes 12: 106 [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793 [DOI] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Ikoma Y, Kato M, Kuniga T, Nakajima N, Yoshida T (2007) Quantification of carotenoids in citrus fruit by LC-MS and comparison of patterns of seasonal changes for carotenoids among citrus varieties. J Agric Food Chem 55: 2356–2368 [DOI] [PubMed] [Google Scholar]
- Mendes AF, Chen C, Gmitter FG Jr, Moore GA, Costa MG (2011) Expression and phylogenetic analysis of two new lycopene β-cyclases from Citrus paradisi. Physiol Plant 141: 1–10 [DOI] [PubMed] [Google Scholar]
- Moise AR, Al-Babili S, Wurtzel ET (2014) Mechanistic aspects of carotenoid biosynthesis. Chem Rev 114: 164–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JC, Cerda A, Simpson K, Lopez-Diaz I, Carrera E, Handford M, Stange C (2016) Increased Nicotiana tabacum fitness through positive regulation of carotenoid, gibberellin and chlorophyll pathways promoted by Daucus carota lycopene β-cyclase (Dclcyb1) expression. J Exp Bot 67: 2325–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8: 68–82 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR (1997) The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA 94: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Liu Q, Yun Z, Guan R, Zeng W, Xu Q, Deng X (2009) Comparative proteomics of a lycopene-accumulating mutant reveals the important role of oxidative stress on carotenogenesis in sweet orange (Citrus sinensis [L.] Osbeck). Proteomics 9: 5455–5470 [DOI] [PubMed] [Google Scholar]
- Pecker I, Gabbay R, Cunningham FX Jr, Hirschberg J (1996) Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals a decrease in its expression during fruit ripening. Plant Mol Biol 30: 807–819 [DOI] [PubMed] [Google Scholar]
- Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Alquézar B, Alós E, Lado J, Zacarías L (2013) Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci Hortic (Amsterdam) 163: 46–62 [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen G, Cohen M, Zamir D, Hirschberg J (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J 17: 341–351 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Shumbe L, Bott R, Havaux M (2014) Dihydroactinidiolide, a high light-induced β-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Mol Plant 7: 1248–1251 [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Schwartz SH, Auldridge M, Taylor MG, Klee HJ (2004) The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J 40: 882–892 [DOI] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Sun L, Yuan B, Zhang M, Wang L, Cui M, Wang Q, Leng P (2012) Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit. J Exp Bot 63: 3097–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, Caporali E, Colombo L, Rotino GL, Trainotti L, Casadoro G (2009) A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. J Exp Bot 60: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N, Hu Z, Liu Q, Xu J, Cheng Y, Guo L, Guo W, Deng X (2007) Expression of phytoene synthase gene (Psy) is enhanced during fruit ripening of Cara Cara navel orange (Citrus sinensis Osbeck). Plant Cell Rep 26: 837–843 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodríguez-Concepción M (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Vainstein A (1999) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4: 232–235 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang S, Lu G, Hou Z, Luo Z, Wang T, Li H, Zhang J, Ye Z (2014) Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J Exp Bot 65: 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P (2007) Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CJ, Fraser PD, Wang WJ, Bramley PM (2006) Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J Agric Food Chem 54: 5474–5481 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene Ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhang J, Nageswaran D, Li L (2015) Carotenoid metabolism and regulation in horticultural crops. Hortic Res 2: 15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma G, Shirai Y, Kato M, Yamawaki K, Ikoma Y, Matsumoto H (2012) Expression and functional analysis of two lycopene β-cyclases from citrus fruits. Planta 236: 1315–1325 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Luo T, Liu C, Wang Y, Yang H, Yang W, Zheng L, Xiao X, Zhang M, Xu R, et al. (2017) An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol 216: 178–192 [DOI] [PubMed] [Google Scholar]