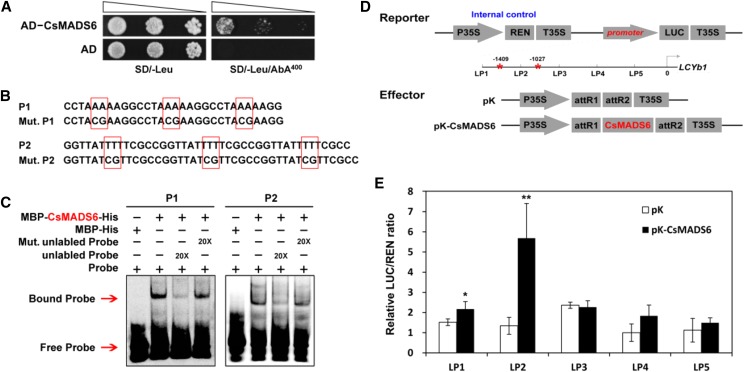
Figure 4.
Interaction of the CsMADS6 protein with the promoter of LCYb1. A, Y1H assay showing the binding of CsMADS6 to the LCYb1 promoter. AD, Empty vector used as the negative control; AD-CsMADS6, prey vector containing CsMADS6; SD/-Leu, SD medium without Leu; SD/-Leu/AbA400, SD medium without Leu supplemented with AbA at the concentration of 400 ng mL−1. Transformed yeast cells were dotted at 10−1 dilutions on the selective medium. B, Probes used for EMSA. P1 and P2, Two Cy5-labeled probes containing three copies of the CArG elements present in the LCYb1 promoter; Mut, P1 and P2 mutant probes (the mutated sites are surrounded by red boxes). C, EMSA showing the binding of CsMADS6 to the LCYb1 promoter. + and − indicate the presence and absence of the indicated probe or protein; 20× indicates a 20-fold excess of unlabeled probe or mutant unlabeled probe. Red arrows indicate the positions of protein-DNA complexes or free probes. D, Schematic diagrams of vectors used for the dual-luciferase assay. The reporter vector contained different promoter fragments of LCYb1 fused to LUC. Red asterisks indicate the positions of the two CArG elements. pK, Empty vector; pK-CsMADS6, overexpression vector containing CsMADS6. E, Dual-luciferase assay showing relative CsMADS6 activation of different promoter fragments of LCYb1. The data are expressed as means ± sd from at least four biological replicates. Asterisks indicate significant differences relative to the empty vector control by one-way ANOVA tests (*, P < 0.05 and **, P < 0.01).