Figure 3. X-ray crystallographic structure of F1Fo ATP synthase illustrating the polyphenol binding pocket.
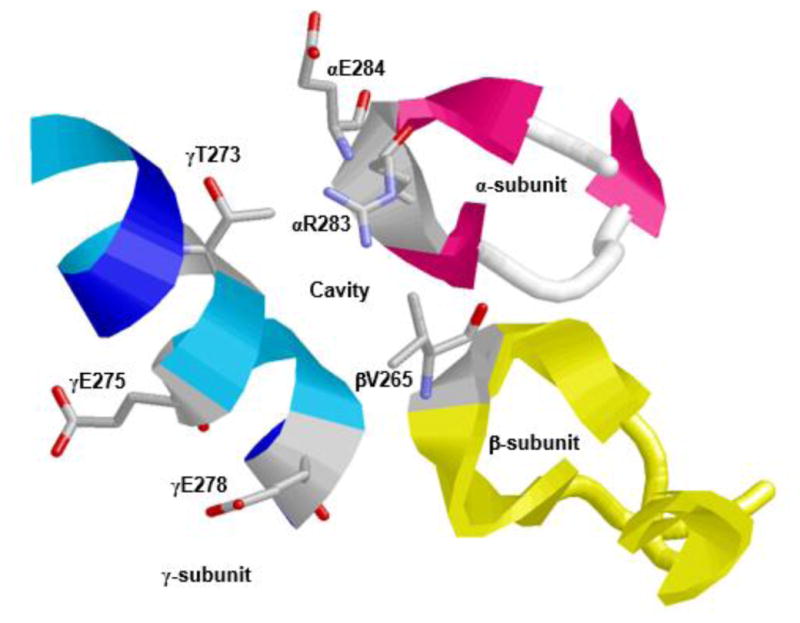
α-, β-, and γ-subunits forming an olive phenolic binding cavity is depicted. Residues αArg-283, αGlu-284, βVal-265, and γThr-273 along with γGln-275 and γGln 278 are identified. RasMol software was used to generate the figure with PDB file 2JJ1 [12]. Escherichia coli residue numbering is used.
Abbreviation: ATP, adenosine triphosphate.
