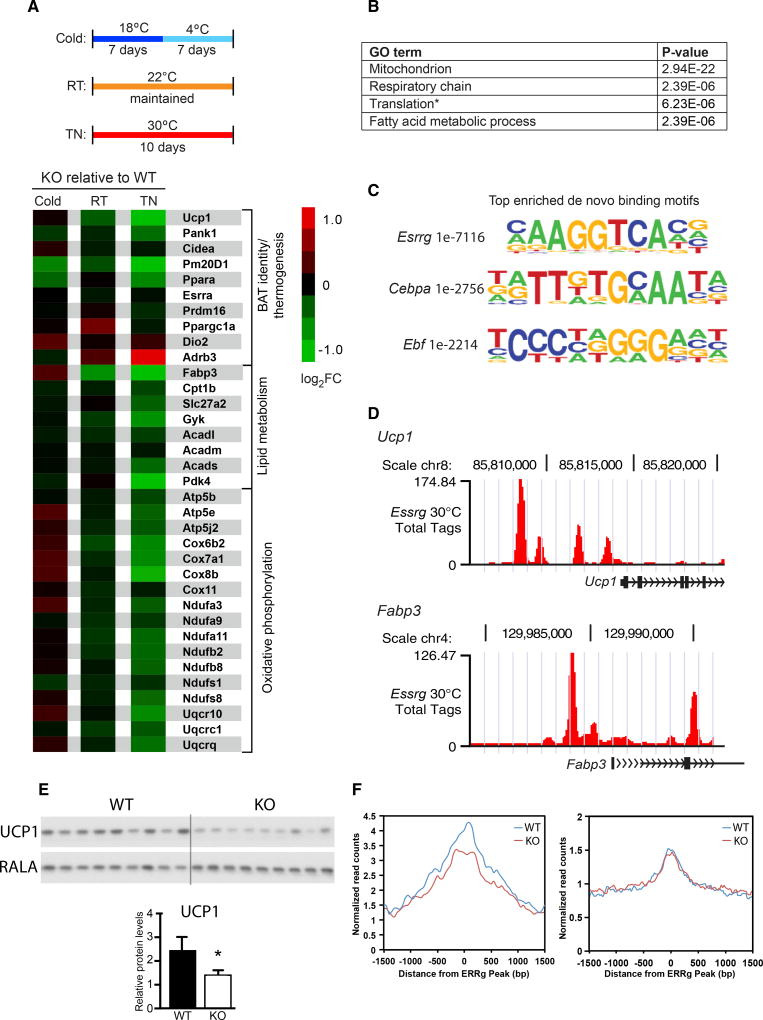
Figure 2. ERRγ Maintains Thermogenic Gene Expression at Thermoneutrality.
(A) Top: schematic of acclimation protocol for flox/flox (WT) and ERRγKO (KO) mice housed under thermoneutral (TN; 30°C), room temperature (RT; 22°C), and cold (18°C, 4°C) conditions. Bottom: heatmap of selected BAT signature genes in BAT from KO relative to WT mice.
(B) Gene Ontology (GO) of enriched pathways or processes in significantly downregulated genes in KO BAT at TN.
(C) Top enriched de novo binding motifs from ERRγ ChIP-seq in BAT of WT mice acclimated to TN.
(D) ERRγ occupancy at the promoters of Ucp1 and Fabp3 in BAT from WT mice acclimated to TN.
(E) Western blot (top) and quantitation (bottom) for UCP1 from WT and KO BAT of mice housed at TN. RALA was used as a loading control.
(F) Average ATAC-seq signals at ERRγ binding sites within ±5 kb of promoters of ERRγ-regulated (left) and non-regulated genes (right) in BAT isolated from WT and ERRγKO mice.
Data represent mean ± SEM. *p < 0.05, Student’s unpaired t test. See also Figure S2.