Abstract
Epithelial sodium channels (ENaC) play an important role in re-absorbing excessive luminal fluid by building up an osmotic Na+ gradient across the tight epithelium in the airway, the lung, the kidney, and the colon. The ENaC is a major pathway for retention of salt in kidney too. MicroRNAs (miRs), a group of non-coding RNAs that regulate gene expression at the post-transcriptional level, have emerged as a novel class of regulators for ENaC. Given the ENaC pathway is crucial for maintaining fluid homeostasis in the lung and the kidney and other cavities, we summarized the cross-talk between ENaC and miRs and recapitulated the underlying regulatory factors, including aldosterone, transforming growth factor-β1, and vascular endothelial growth factor-A in the lung and other epithelial tissues/organs. We have compared the profiling of miRs between normal and injured mice and human lungs, which showed a significant alteration in numerous miRs in mouse models of LPS and ventilator induced ARDS. In addition, we reiterated the potential regulation of the ENaC by miRs in stem/progenitor cell-based re-epithelialization, and identified a promising pharmaceutic target of ENaC for removing edema fluid in ARDS by mesenchymal stem cells-released paracrine. In conclusion, it seems that the interactions between miRs and scnn1s/ENaCs are critical for lung development, epithelial cell turnover in adult lungs, and re-epithelialization for repair.
Keywords: Epithelial reparation, Stem/progenitor cells, Re-epithelialization, Non-coding RNA, Transepithelial sodium transport, Cross talk, Amiloride-sensitive sodium channels
Introduction
Non-coding microRNAs (miRs) emerged as a major class of gene expression regulators in general, which belong to single-stranded RNA segments with approximately 20–24 nucleotides in length. It is proposed that miRs play a crucial role in the regulation of ion channels, as implicated in dysregulation of ion channels and restoring or downregulating the expression levels of specific miRs could be a promising approach in channelopathies [1, 2]. For example, KCNJ2 gene, which encodes the pore-forming α-subunit Kir2.1 as a main K+ channel subunit of IK1, was a target of miR-1. miR-1 regulated cardiac conductions and contributed to ischemic arrhythmias in myocardial infarction via targeting Kir2.1 as well as connexin 43. Repression of KCNJ2/Kir2.1/IK1 by miR-1 might mediate the slowing down of cardiac conduction following a shift of the membrane potential to depolarized levels to inactivate Na+ channels [3]. In lung epithelial cells, miR-16, miR-101/miR-144, and miR-96/miR-330 specifically bound to the 3′-untranslated region (3′-UTR) of the β subunit of epithelial sodium channels (ENaC) [4], cystic fibrosis transmembrane conductance regulator (CFTR) [5], and aquaporin 5 [6], respectively.
The apically located ENaC proteins, including the α, β, γ, and δ subunits, are encoded by the scnn1a, scnn1b, scnn1g, and scnn1d genes, respectively. These ENaC genes are expressed in the alveolar, renal, colon, and airway epithelial cells, and serve as a critical pathway for maintaining luminal fluid homeostasis in these tissues/organs. They are crucial for resolution of fluid in edematous diseases. Several factors have been found to regulate ENaC expression and/or function in alveolar epithelial cells, including aldosterone, growth factors, protein kinases, and serine proteases. The corresponding mechanisms have been reviewed previously [7–10]. As transmembrane proteins are capable of sensing both intracellular and extracellular signals, ion channels have been proposed to be candidate epigenetic regulators linking extracellular signals to miRs during the morphogenesis of fetal organs [11]. This is supported by an innovative study of embryo implantation that scnn1s/ENaCs regulated miR-101/miR-199a-3p via cAMP-response element binding protein (CREB) phosphorylation [12]. Using miRNA arrays, Zhang and co-workers reported that scnn1a/αENaC was a potential target of miR-125b to up-regulate the replication of hepatitis B virus in HepG2.2.15 cells [13]. It is emerging that scnn1s/ENaCs may serve as both up and downstream signal molecules of miR-initiated pathways. In addition, ENaCs may indirectly regulate miR-mediated cell processes, as ENaC-interactive CFTR transduced the extracellular bicarbonate signal into activation of miR-125b to target the p53 pathway [14].
The purpose of this review is to identify and systematically explore the dynamically regulated miRs and potential miR-ENaC interactive networks specific to the transition from amniotic fluid-breathing to air-breathing lungs as well as lung injury and regeneration. To date, a considerable progress has been made in the studies of the interactions between miRs and the expression and function of ENaC genes. In addition, stem/progenitor cells release exosomes/microvesicles composed of multiple miRs. These miRs may involve in the benefits of stem/progenitor cells in re-alveolarization of injured lungs through targeting the ENaC signaling. However, this emerging field has not been summarized to date. Herein we review the accumulated evidence regarding the alterations in critical lung-specific miRs in injured lungs, the cross-talks between miRs and scnn1s/ENaCs, and the potential roles of miRs in stem/progenitor cell-induced reparative processes of injured respiratory epithelium.
Biogenesis and nomenclature of miRs
MiRs are a category of non-coding single-stranded RNA segments that are encoded by endogenous genes [12]. Through complete or incomplete pairing with the complementary sequences of 3′-UTR of targeting mRNAs, miRs degrade the 3′-UTR chain or interrupt translation [15, 16]. Apparently, miRs regulate both transcriptional and translational processes either by increasing mRNA degradation or inhibiting protein translation [17, 18], and thus negatively alter the synthesis of the corresponding proteins and ultimately regulating multiple cellular activities [19–22]. For example, conditional deletion of dicer in embryonic lung epithelium led to abnormal branching and increased apoptosis, indicating that miRs may have crucial roles in lung development [23].
To date, 1, 881 hairpin precursors and 2, 588 mature miR sequences have been identified in humans; while 1, 193 hairpin precursors and 1, 915 mature miR sequences in mice. Of note, many could remain to be discovered about their functional targets and biologic roles (miRBase online server, http://www.mirbase.org/index.shtml).
Primary miRNA transcripts (pri-miRs) are produced in the nucleus by RNA polymerase II and at the same time some pri-miRs can also be produced by RNA polymerase III [24]. Subsequently pri-miRs are processed into pre-miRs, which are exported to the cytoplasm finally resulting in mature miRs [25]. Mature miRs trigger down-regulation and suppression of target gene expressions via binding target mRNAs at complementary sequences in 3′-UTRs or coding sequences [19, 26].. A single miR may regulate the expression of multiple genes, and multiple miRs may modulate the same target gene. Furthermore, the expression of miRs showed both tissue/cell specificity and spatial/temporal specificity [27].
The nomenclature guidelines of miRs generally follow the four basic rules: 1) The abbreviation of species is as a prefix, such as Homo sapiens (hsa), Mus musculus (mmu). The mature sequences are designated ‘miR’ in the database, whereas the precursor hairpins are labelled ‘mir’. 2) The miRNAs is abbreviated as miRs, according to its cloned successively and continuous serial number. 3) The gene names are intended to convey limited information about functional relationships between mature miRs. The homologous miRs of different species have the same sequence so use the same name. 4) For chromosome transcription processing together with the same miRs sequence, but with different pre-mature miR sequence, in order to distinguish, mature miRs have numbered suffixes.
Development-dependent profile of lung miRs and ENaC
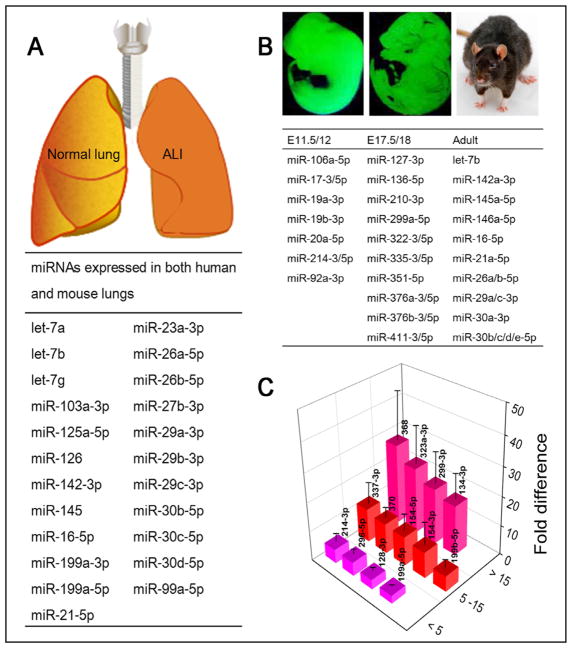
The relative levels of all miRs expressed in adult mouse and human lungs were quantitatively analyzed by a RT-PCR based approach. The top 30 miRs predominately expressed in human and mouse lungs were confirmed [28]. The data identified a substantial similar profile between two species, for 23 of these predominately expressed miRs were shared by both mouse and human (Fig. 1A). Of note, miR-26a was the most highly expressed individual miR, whilst the let-7 family, miR-29 family, miR-30 family, and miR-199 were also expressed highly in both species. These qPCR data were further validated by other groups [29–32].
Fig. 1.
Species- and development-dependent miR expression in the lungs. A. Predominant expressed 23 miRs in both mouse and human lungs [28]. B. Development-dependent miR profiles. MiRs have the highest expression in the lungs of early embryonic (E11.5/12), the end of the canalicular stages (E17.5/18), and adults. C. miR expression difference between fetal and adult human lungs. Data are adapted from Williams AE et al [28].
Multiple miRs were shown to be differentially expressed during lung development. The evidence that miRs play a role in lung development derives from mice with conditioning knockout of dicer in lung epithelial cells [23]. These mice exhibited a failure of epithelial branching, underscoring the essential regulatory role of miRs in lung epithelial morphogenesis [23, 33]. To determine if miRs have an evolutionary conserved role in lung development, two studies found that 117 and 198 miRs were dynamically altered during mouse lung organogenesis, respectively [34, 35]. Fig. 1B shows the miRs expressed at the highest level at different stages of lung development. miR microarrays were carried out for total RNA from lungs at different embryonic stages and the adulthood. About 37% (73/198) of miRs had no or very low expression in each group. By contrast, 15 miRs showed a peak at E17.5/18, the canalicular stage.
The altered expression of both miRs and ENaC subunits before and after birth may be related to the fast resolution of amniotic fluid from the air spaces at birth. The development-dependent expression of both ENaC and miRs in the human respiratory system was confirmed [28, 36]. The β and γ ENaC subunits were not detected in human lungs at the early stage of embryonic development [36]. Near before birth, increased ENaC activity at the apical surface of lung epithelial cells was observed [37]. In the adult lungs, all of four subunits (α, β, γ and δ) were detected [38, 39]. It has been speculated that the increased ENaC activity was responsible for resolving amniotic fluid in the air spaces in neonates [40]. This notion was supported by the association between depressed ENaC expression and respiratory distress syndrome in newborns or animals deficient in scnn1 genes [41]. Evidence has shown that a tumor suppressor, Rbl2 which belongs to the Rb family, as a new target in promoting the high proliferation and undifferentiated phenotype of lung epithelial progenitor cells [35]. And the regulation of ENaC by ERK, PKC, cAMP, PKA, and TNF may play a role in the differentiation of endogenous progenitor/stem cells [42]. However, the mechanisms for the changes of ENaC and miRs from embryonic to alveolar phases during lung development and their potential correlations have not been fully known yet. During the transition of fluid-filled lungs to the first breath, the marked differences in lung miRs prior to and post birth would provide critical clue for identifying miRs specifically associating with ENaC function.
Williams AE and co-workers compared the expression profiles of miRs between fetal and adult human lungs [28]. In this case, 13 miRs were expressed at a greater level in fetal lungs than adult controls (Fig. 1C). In particular, the levels of miR-134-3p, miR-299-3p, miR-323a-3p, and miR-368 in human fetal lungs were more than 15-fold than those in adult preparations. Diversely, expression of three miR-29 family members (a, b, and c) ranked the highest in adult. Significantly, many of these striking alterations in birth-induced miR expression were common in mice. The expression of miR-134, miR-214, miR-296, miR-299, miR-323, miR-337 and miR-370 was highly expressed in both mouse and human fetal lungs and subsequently down-regulated in adult lungs.
Bhaskaran M et al’s classic studies systematically characterized rat lung miR profiles between full-term gestation (E21) and neonatal rats. Compared with fetal lungs, cluster 4 miRs, including miR-18, miR-127, and miR-351 dramatically reduced on day 0 (P0) and continuously declined on day 6 (P6) [43]. In sharp contrast, miR-210 showed a significantly increase on P0 followed by a reduction on P6. The changes in lung miRs could be associated with the abundance of ENaC expression and resolution of amniotic fluid from the air spaces within 48 h after birth.
Inter-regulation of miRs and ENaCs in normal and diseased tissues
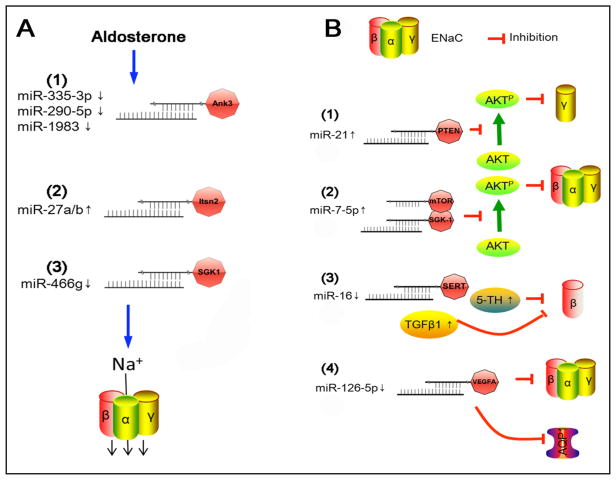
A significant reduction in the expression of miR-335-3p, miR-290-5p, and miR-1983 was proposed to lead to an increment in ENaC-mediated sodium transport by aldosterone in mouse cortical collecting duct (CCD) cells [44]. In contrast, aldosterone up regulated expression of miR-27 [45]. Even in the absence of aldosterone, the miR-27 increased ENaC activity in CCD cells. Furthermore, mimic miR-27a/b was verified to bind to the 3′-UTR of intersectin-2 to depress endocytosis of ENaC proteins. miR-466g, as a novel aldosterone responsive molecule, regulated ENaC activity in CCD cells probably through the glucocorticoid regulated kinase 1 pathway [46]. These miRs reinforced an important role for miRs as a new element in the renin–angiotensin–aldosterone system signaling cascade that maintains Na+ transport in the kidney (Fig. 2A).
Fig. 2.
ENaC-targeting miRs and signal pathways in renal and lung epithelial cells. A. Aldosterone responsive miRs targeting ENaC in CCD cells. B. Mechanisms for lung miRs to regulate ENaC.
In addition to be a downstream molecule of the signal axis, ENaCs might serve as regulators of miRs in different physiological contexts [12]. For example, proteolytically cleaved by embryo implantation-released serine protease, ENaCs depressed the expression of miR-101 and miR-199a-3p, most likely through CREB phosphorylation in endometrial epithelial cells [12]. Consequently, phosphorylation of CREB and up-regulation of cyclooxygenase type 2, facilitate prostaglandin production and release required for embryo implantation [47].
Acute respiratory distress syndrome (ARDS) is a clinical syndrome of acute respiratory characterized by protein-rich pulmonary edema. Removal of edema fluid from the air spaces via the ENaC pathway in ARDS is crucial for resolution of ARDS. The main pathological characteristics of ARDS include an increase in the permeability of the blood-gas barrier, flooding of alveoli, and deposition of serum proteins into the alveoli. By comparing the miR profiles between normal and injured lungs, a significant alteration in numerous miRs was reported in mouse models of LPS and ventilator induced ARDS [31, 32, 48–54]. The expression of lung miR-7, miR-21, miR-34, miR-124, miR-125, miR-129, and miR-205 were increased, whereas lung miR-16 decreased significantly. Among them, miR-16 and miR-21 were predominately expressed in the human and mouse lungs. Of note, these miRs were predicted to target the sequences of the 3′-UTR of ENaC subunits. (Fig. 2B).
Four groups of ENaC-targeting miRs have been identified in ARDS models to date. These potentially therapeutic miR targets for improving ENaC function in ARDS are summarized in Fig. 2B. 1) Lung and serum miR-21 concentration increased significantly in ARDS patients. miR-12 reduced the expression of γ ENaC both at the protein and mRNA levels via the PTEN/AKT signaling pathway in LPS-induced rat ARDS model. It is possible that elimination of the increment in miR-21 could represent a novel target to restore ENaC activity in ARDS [55]. 2) A549 epithelial cells have some characteristics of authentic alveolar epithelial type 2 cells and when challenged by LPS, there was a reduction in the expression of miR-7-5p restored the expression of αβγ ENaC protein via silencing both the target of rapamycin and glucocorticoid regulated kinase 1 genes. It may be feasible that ENaC expression and function could be benefited by a reduction in miR-7-5p expression [15]. 3) Although diverse results had been reported, there was a significant decrease in lung β ENaC in hyperoxic mice compared to normoxic controls. Concurrently, a marked reduction in miR-16 was identified. Moreover, miR-16 could up regulate the expression of β ENaC protein by suppressing the release of transforming growth factor-β1, a critical inhibitory signaling pathway for ENaC [4]. 4) miR-126-5p ameliorated dysfunctional alveolar fluid clearance probably by down regulating vascular endothelial growth factor-A, in turn, maintaining the activity of αβγ ENaC in alveolar epithelial type 2 (AT2) cells [56].
Pulmonary stem/progenitor cell-released miRs for injury repair
Several lineages of stem/progenitor cells expressing scnn1s/ENaCs have been identified in the airways and the lungs [42]. These stem/progenitor cells include basal cells, Clara/club cells, bronchioalveolar stem cell (BASC), AT2 cells, and side population cells [57–60]. Increasing evidence from developmental biology and regenerative medicine demonstrates that these cells contribute to morphogenesis during embryonic development, turnover in adult lungs, and regeneration of injured airway and alveolar tissues. miRs were implicated in the lung development associated with stem cells [61, 62]. These observations have further been confirmed by two classic studies demonstrating that miRs participated in a series of biological processes mediated by stem cells, ranging from embryogenesis to programmed cell death [63, 64].
BASCs possessed a unique miR profile [57]. The microarrays identified that 56 miRs were up-regulated in BASCs. On the other hand, 60 miRs were down-regulated, i.e., miR-142-3p, miR-451, miR-106a, miR-142-5p, miR-15b, miR-20a, miR-106b, miR-25, and miR-486, compared to control cells (CD31neg CD45negCD34negSca-1neg). These observations suggest that miRs might have important roles in maintaining the self-renewal capacity of BASCs. Dysregulated expression could be involved in the conversion of BASCs into lung cancer stem cells [65]. Additionally, miR-17-92 cluster promoted the high proliferation and undifferentiated phenotype of embryonic lung epithelium that over-expresses surfactant protein C, which was highly expressed in embryonic stem cells and declined during mouse lung development [35]. This miR cluster is a conserved gene encoding 7 miRs (miR-17-5p, miR-17-3p, miR-18, miR-19a, miR-20a, miR-19b-1, and miR-92-1).
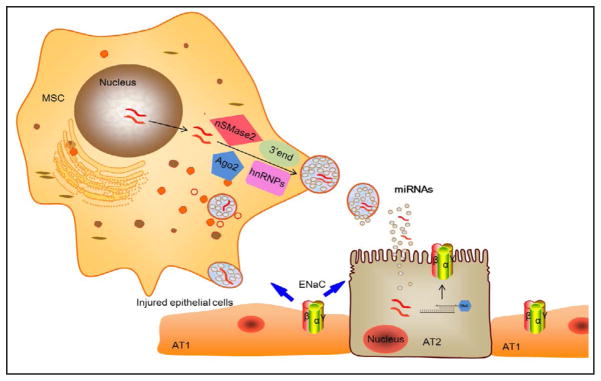
Microvesicles (MVs) have been described as an important mechanism of intercellular communication [66]. MVs are the plasma membrane derived vesicles/exosomes released by many cell types [67, 68], including lung stem and progenitor cells [69–72]. MVs, as one of paracrine mechanisms, are a major pathway for stem cells to rescue adjunct damaged cells [73, 74]. MVs contain a population of mRNAs and miRs of 17–22nt in addition to a distinctive set of proteins [68, 72, 75–80]. Stem/progenitor cells released miRs to stimulate themselves to proliferate and later differentiate into functional cells to replace injured cells. miR-containing MVs could be delivered to injured cells via paracrine resulting in an alteration of target cell behavior [76]. miRs were detected in both MSCs and conditional medium with microarray. MVs secreted by MSCs contained 60 miRs: 15 miRs were only in conditional medium, and 12 miRs were detected in conditional medium 2 fold than those in MSCs [72]. Intriguingly, some of the miRs were only found in MVs or concentrated in the MVs at high levels, suggesting that the miRs may be uniquely packed into MVs to be shuttled into adjunct cells with multiple functions. Additionally, MSCs secreted MVs were enriched in pre-miRs too [72]. MVs derived from embryonic stem cells contained abundant miRs that could be transferred to mouse embryonic fibroblasts in vitro [80]. In view of the extensive regulatory capacity of miRs that mediate posttranscriptional control of gene expression [81, 82], stem cells may therefore modulate the biological functions by delivering genetic information and altering the gene expression of target cells.
There is evidence that overexpression of miR-183 directly targeted the expressions of human α, β, γ ENaC subunits in stem cells [83]. Drosophila miR-263a, the ortholog of human miR-183, reduced the expression of ENaC subunits in enterocytes. Deletion of miR-263a dehydrated intraluminal surface of the intestinal track and increased enterocytosolic Na+ content, a cystic fibrosis-like phenotype with increased bacterial load and expression of antimicrobial peptides. This study linked ENaC-targeting miRs to stem cell homeostasis via controlling proliferation and differentiation in Drosophila.
Stem cell-based therapy is a promising therapeutic approach for ARDS. Much of current interests of MSCs have focused on soluble factors due to their ability to secrete paracrine, including miRs [84, 85], which regulate endothelial and epithelial permeability [86–88], restore alveolar fluid clearance [89], exhibit immunosuppression [90–92], normalize dysregulated inflammation [93, 94], enhance tissue repair, and inhibit bacterial growth [95]. In particular, ENaC may be a major target for removing edema fluid by MSC-released paracrine in the ARDS lungs (Fig. 3). Intriguingly, ENaC serves as a regulator to maintain intestinal stem cell homeostasis through cell swelling-activated JNK signal pathways [83]. In addition, ENaC contributed to beneficiary effects of KGF and other paracrine for injured lungs.
Fig. 3.
Potential inter-regulation of MSC-released miRNAs and ENaC subunits in re-alveolarization. MSCs as well as AT2 cells transfer the cytosolic materials in microvesicles to adjunct injured AT1 cells. miRs released by microvesicles would target the expression and activity of ENaCs, which in turn, regulate stem/progenitor cell homeostasis by altering proliferation and differentiation.
In addition to cell-based therapy, the effects of miRs and MVs for ARDS have drawn increasing attention in recent years [96]. As critical regulators of gene expression, miRs regulate numerous developmental, physiological, and pathogenic processes, including lung development, ARDS, asthma, and lung cancer [97]. Targeted deletion of dicer, led to embryonic lung abnormal development manifested by apoptosis and abnormal branch of the airway [98, 99]. Although the preclinical evidence for MV and miR-based therapy is accumulating rapidly and promising, the relationships between MSC-released miRs and ENaC are unclear.
Conclusion
This review summarized the lung-specific miRs, compared the expression profiles between fetal and adult lungs, recapitulated the development dependence and predominant miRs in adult lungs, the ENaC-targeting miRs in ARDS lungs, and the interactions between ENaC subunits and miRs in stem/progenitor cell-based re-epithelialization. The cross-talk between scnn1s/ENaCs and miRs may lead to the development of new strategies for ARDS by manipulating the expression of lung ENaCs and/or delivery of miRs. At present, miRs open novel fields in biomarker research, diagnosis, and therapy. Restoring or downregulating the expression levels of specific miRs could be a promising approach and miRNA-based therapeutics could be a remarkably smart way for targeting therapy in cancer and other related diseases. However, most studies so far have been performed in vitro, in vivo studies based on systemic or localized target delivery of miRNA mimics or antagomirs are still required to be carried out. In contrast to cytosolic miRs, the inter-regulation of miRs and scnn1s/ENaCs in mitochondria and other cellular organelles is unknown. MiR-21 was targeted as a clinical trial for lung cancer and ENaC modulators were tested for ARDS patients previously [100]. To date, the benefits of targeting ENaC-interactive miRs have not been investigated yet.
To study the interactions of miRs and ENaC subunits, in silico prediction is required for screening potential binding sites in scnn1s/ENaCs. The algorithms of computational prediction for miRs to target ENaC subunits have been developed based on common features of known miRs and their mRNA target interactions [21, 101]. Unfortunately, the predicted results between these prediction servers, including Miranda and DIANA-microT, are inconsistent, most likely due to diverse algorithms. A more precise software with combined algorithms and validated with experimental data is needed. This may require more preclinical studies to identify the true binding sites by bench works. In addition, current computational prediction tools are diverse, both in approach and performance. It is difficult to decide which predicted miR-ENaC interactions are more likely to be accurate, and which tool provides the best performance [21, 101].
In conclusion, it seems that the interactions between miRs and scnn1s/ENaCs are critical for lung development, epithelial cell turnover, and lung repair post injury. However, this emerging topic is still in its early stage. There are several unanswered questions, including the beneficial effects of individual miR, the mechanisms and signaling pathways, and therapeutic implications for ARDS.
Acknowledgments
This work was supported by grants from the American Heart Association (AHA 14GRNT20130034 and 16GRNT30780002), National Institute of Health (NIH HL134828, HL116826, AI133465, and HL51856), the National Natural Science Foundation of China (NSFC 81670010), and Basic Research Project of Liaoning Higher School (LQNK201745).
Abbreviations
- miRs
Non-coding microRNAs
- ENaC
epithelial sodium channels
- CFTR
cystic fibrosis transmembrane conductance regulator
- CREB
response element binding protein
- 3′-UTR
3′-untranslated region
- pri-miRs
Primary miRNA transcripts
- has
Homo sapiens
- mmu
Mus musculus
- CCD
cortical collecting duct
- ARDS
Acute respiratory distress syndrome
- BASC
bronchioalveolar stem cell
- MVs
Microvesicles
- MSC
mesenchymal stem cells
- Ank3
ankyrin3
- Itsn2
intersectin-2
- SGK1
serum and glucocorticoid regulated kinase 1
- PTEN
phosphatase and tensin homolog
- AKT
protein kinase B
- AKTP
phosphorylation of protein kinase B
- mTOR
mammalian target of rapamycin
- SERT
serotonin transporter
- 5-TH
serotonin
- VEGFA
vascular endothelial growth factor-A
- TGF β1
transforming growth factor β1
- AQP1
aquaporin 1
- AT2
alveolar epithelial type 2
- nSMase2
neutral sphyngomyelinase 2
- hnRNPA2B1
heterogeneous nuclear ribonucleoprotein A2B1
- Ago2
argonaute2
Footnotes
Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Elvira-Matelot E, Jeunemaitre X, Hadchouel J. Regulation of ion transport by microRNAs. Curr Opin Nephrol Hypertens. 2011;20:541–546. doi: 10.1097/MNH.0b013e328348b4aa. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z. miRNA in the regulation of ion channel/transporter expression. Compr Physiol. 2013;3:599–653. doi: 10.1002/cphy.c110002. [DOI] [PubMed] [Google Scholar]
- 3.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 4.Tamarapu Parthasarathy P, Galam L, Huynh B, Yunus A, Abuelenen T, Castillo A, Kollongod Ramanathan G, Cox R, Jr, Kolliputi N. MicroRNA 16 modulates epithelial sodium channel in human alveolar epithelial cells. Biochem Biophys Res Commun. 2012;426:203–208. doi: 10.1016/j.bbrc.2012.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan F, Nuovo GJ, Crawford M, Boyaka PN, Kirkby S, Nana-Sinkam SP, Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Chen M, Zhang Y, Peng P, Li J, Xin X. miR-96 and miR-330 overexpressed and targeted AQP5 in lipopolysaccharide-induced rat lung damage of disseminated intravascular coagulation. Blood Coagul Fibrinolysis. 2014;25:731–737. doi: 10.1097/MBC.0000000000000133. [DOI] [PubMed] [Google Scholar]
- 7.Verrey F, Loffing J, Zecevic M, Heitzmann D, Staub O. SGK1: aldosterone-induced relay of Na+ transport regulation in distal kidney nephron cells. Cell Physiol Biochem. 2003;13:21–28. doi: 10.1159/000070246. [DOI] [PubMed] [Google Scholar]
- 8.Rossier BC, Stutts MJ. Activation of the epithelial sodium channel (ENaC) by serine proteases. Annu Rev Physiol. 2009;71:361–379. doi: 10.1146/annurev.physiol.010908.163108. [DOI] [PubMed] [Google Scholar]
- 9.Eaton DC, Helms MN, Koval M, Bao HF, Jain L. The contribution of epithelial sodium channels to alveolar function in health and disease. Annu Rev Physiol. 2009;71:403–423. doi: 10.1146/annurev.physiol.010908.163250. [DOI] [PubMed] [Google Scholar]
- 10.Hanukoglu I, Hanukoglu A. Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene. 2016;579:95–132. doi: 10.1016/j.gene.2015.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Zhang JT, Chan HC. Ion channels/transporters as epigenetic regulators?-a microRNA perspective. Sci China Life Sci. 2012;55:753–760. doi: 10.1007/s11427-012-4369-9. [DOI] [PubMed] [Google Scholar]
- 12.Sun X, Ruan YC, Guo J, Chen H, Tsang LL, Zhang X, Jiang X, Chan HC. Regulation of miR-101/miR-199a-3p by the epithelial sodium channel during embryo implantation: involvement of CREB phosphorylation. Reproduction. 2014;148:559–568. doi: 10.1530/REP-14-0386. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Chen J, He Y, Zhan X, Zhao R, Huang Y, Xu H, Zhu Z, Liu Q. miR-125b inhibits hepatitis B virus expression in vitro through targeting of the SCNN1A gene. Arch Virol. 2014;159:3335–3343. doi: 10.1007/s00705-014-2208-y. [DOI] [PubMed] [Google Scholar]
- 14.Lu YC, Chen H, Fok KL, Tsang LL, Yu MK, Zhang XH, Chen J, Jiang X, Chung YW, Ma AC, Leung AY, Huang HF, Chan HC. CFTR mediates bicarbonate-dependent activation of miR-125b in preimplantation embryo development. Cell Res. 2012;22:1453–1466. doi: 10.1038/cr.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin K, Zhong X, Wang D. MicroRNA-7-5p regulates human alveolar epithelial sodium channels by targeting the mTORC2/SGK-1 signaling pathway. Exp Lung Res. 2016;42:237–244. doi: 10.1080/01902148.2016.1197347. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 23.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 25.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 27.Staszel T, Zapala B, Polus A, Sadakierska-Chudy A, Kiec-Wilk B, Stepien E, Wybranska I, Chojnacka M, Dembinska-Kiec A. Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn. 2011;121:361–366. [PubMed] [Google Scholar]
- 28.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaporidi K, Vergadi E, Kaniaris E, Hatziapostolou M, Lagoudaki E, Georgopoulos D, Zapol WM, Bloch KD, Iliopoulos D. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;303:L199–207. doi: 10.1152/ajplung.00370.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J, Jiang G, Asmann YW, Tomaszek S, Jen J, Kislinger T, Wigle DA. MicroRNA networks in mouse lung organogenesis. PLoS One. 2010;5:e10854. doi: 10.1371/journal.pone.0010854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaillard D, Hinnrasky J, Coscoy S, Hofman P, Matthay MA, Puchelle E, Barbry P. Early expression of beta- and gamma-subunits of epithelial sodium channel during human airway development. Am J Physiol Lung Cell Mol Physiol. 2000;278:L177–184. doi: 10.1152/ajplung.2000.278.1.L177. [DOI] [PubMed] [Google Scholar]
- 37.Matalon S, Bartoszewski R, Collawn JF. Role of epithelial sodium channels in the regulation of lung fluid homeostasis. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1229–1238. doi: 10.1152/ajplung.00319.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji HL, Su XF, Kedar S, Li J, Barbry P, Smith PR, Matalon S, Benos DJ. Delta-subunit confers novel biophysical features to alpha beta gamma-human epithelial sodium channel (ENaC) via a physical interaction. J Biol Chem. 2006;281:8233–8241. doi: 10.1074/jbc.M512293200. [DOI] [PubMed] [Google Scholar]
- 39.Woollhead AM, Sivagnanasundaram J, Kalsi KK, Pucovsky V, Pellatt LJ, Scott JW, Mustard KJ, Hardie DG, Baines DL. Pharmacological activators of AMP-activated protein kinase have different effects on Na+ transport processes across human lung epithelial cells. Br J Pharmacol. 2007;151:1204–1215. doi: 10.1038/sj.bjp.0707343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji HL, Zhao RZ, Chen ZX, Shetty S, Idell S, Matalon S. delta ENaC: a novel divergent amiloride-inhibitable sodium channel. Am J Physiol Lung Cell Mol Physiol. 2012;303:L1013–1026. doi: 10.1152/ajplung.00206.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Koshy S, Folkesson HG. RNA interference for CFTR attenuates lung fluid absorption at birth in rats. Respir Res. 2008;9:55. doi: 10.1186/1465-9921-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Jiang BJ, Zhao RZ, Ji HL. Epithelial sodium channels in pulmonary epithelial progenitor and stem cells. Int J Biol Sci. 2016;12:1150–1154. doi: 10.7150/ijbs.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edinger RS, Coronnello C, Bodnar AJ, Labarca M, Bhalla V, LaFramboise WA, Benos PV, Ho J, Johnson JP, Butterworth MB. Aldosterone regulates microRNAs in the cortical collecting duct to alter sodium transport. J Am Soc Nephrol. 2014;25:2445–2457. doi: 10.1681/ASN.2013090931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Edinger RS, Klemens CA, Phua YL, Bodnar AJ, LaFramboise WA, Ho J, Butterworth MB. A microRNA cluster miR-23-24-27 is upregulated by aldosterone in the distal kidney nephron where it alters sodium transport. J Cell Physiol. 2017;232:1306–1317. doi: 10.1002/jcp.25599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs ME, Kathpalia PP, Chen Y, Thomas SV, Noonan EJ, Pao AC. SGK1 regulation by miR-466g in cortical collecting duct cells. Am J Physiol Renal Physiol. 2016;310:F1251–1257. doi: 10.1152/ajprenal.00024.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruan YC, Guo JH, Liu X, Zhang R, Tsang LL, Dong JD, Chen H, Yu MK, Jiang X, Zhang XH, Fok KL, Chung YW, Huang H, Zhou WL, Chan HC. Activation of the epithelial Na+ channel triggers prostaglandin E(2) release and production required for embryo implantation. Nat Med. 2012;18:1112–1117. doi: 10.1038/nm.2771. [DOI] [PubMed] [Google Scholar]
- 48.Zhou T, Garcia JG, Zhang W. Integrating microRNAs into a system biology approach to acute lung injury. Transl Res. 2011;157:180–190. doi: 10.1016/j.trsl.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo ZL, Ren T, Xu L, Zhang L, Yin Q, Wang JC, Liang YJ. The microRNAs expression changes rapidly in mice lung tissue during lipopolysaccharide-induced acute lung injury. Chin Med J (Engl) 2013;126:181–183. [PubMed] [Google Scholar]
- 50.Cao Y, Lyu YI, Tang J, Li Y. MicroRNAs: Novel regulatory molecules in acute lung injury/acute respiratory distress syndrome. Biomed Rep. 2016;4:523–527. doi: 10.3892/br.2016.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Z, Gong H, Li Y, Jie K, Ding C, Shao Q, Liu F, Zhan Y, Nie C, Zhu W, Qian K. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res. 2013;39:275–282. doi: 10.3109/01902148.2013.808285. [DOI] [PubMed] [Google Scholar]
- 52.Cai ZG, Zhang SM, Zhang Y, Zhou YY, Wu HB, Xu XP. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol. 2012;90:37–43. doi: 10.1139/y11-095. [DOI] [PubMed] [Google Scholar]
- 53.Xie T, Liang J, Liu N, Wang Q, Li Y, Noble PW, Jiang D. MicroRNA-127 inhibits lung inflammation by targeting IgG Fcgamma receptor I. J Immunol. 2012;188:2437–2444. doi: 10.4049/jimmunol.1101070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vergadi E, Vaporidi K, Theodorakis EE, Doxaki C, Lagoudaki E, Ieronymaki E, Alexaki VI, Helms M, Kondili E, Soennichsen B, Stathopoulos EN, Margioris AN, Georgopoulos D, Tsatsanis C. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol. 2014;192:394–406. doi: 10.4049/jimmunol.1300959. [DOI] [PubMed] [Google Scholar]
- 55.Qi W, Li H, Cai XH, Gu JQ, Meng J, Xie HQ, Zhang JL, Chen J, Jin XG, Tang Q, Hao Y, Gao Y, Wen AQ, Xue XY, Gao Smith F, Jin SW. Lipoxin A4 activates alveolar epithelial sodium channel gamma via the microRNA-21/PTEN/AKT pathway in lipopolysaccharide-induced inflammatory lung injury. Lab Invest. 2015;95:1258–1268. doi: 10.1038/labinvest.2015.109. [DOI] [PubMed] [Google Scholar]
- 56.Tang R, Pei L, Bai T, Wang J. Down-regulation of microRNA-126-5p contributes to overexpression of VEGFA in lipopolysaccharide-induced acute lung injury. Biotechnol Lett. 2016;38:1277–1284. doi: 10.1007/s10529-016-2107-2. [DOI] [PubMed] [Google Scholar]
- 57.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 58.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffman AM, Ingenito EP. Alveolar epithelial stem and progenitor cells: emerging evidence for their role in lung regeneration. Curr Med Chem. 2012;19:6003–6008. doi: 10.2174/092986712804485872. [DOI] [PubMed] [Google Scholar]
- 60.Ling TY, Liu YL, Huang YK, Gu SY, Chen HK, Ho CC, Tsao PN, Tung YC, Chen HW, Cheng CH, Lin KH, Lin FH. Differentiation of lung stem/progenitor cells into alveolar pneumocytes and induction of angiogenesis within a 3D gelatin--microbubble scaffold. Biomaterials. 2014;35:5660–5669. doi: 10.1016/j.biomaterials.2014.03.074. [DOI] [PubMed] [Google Scholar]
- 61.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 62.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dumortier O, Hinault C, Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312–324. doi: 10.1016/j.cmet.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Qian S, Ding JY, Xie R, An JH, Ao XJ, Zhao ZG, Sun JG, Duan YZ, Chen ZT, Zhu B. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377:668–673. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 66.Hugel B, Martinez MC, Kunzelmann C, Freyssinet JM. Membrane microparticles: two sides of the coin. Physiology (Bethesda) 2005;20:22–27. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 67.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 70.Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 71.Sze SK, de Kleijn DP, Lai RC, Khia Way Tan E, Zhao H, Yeo KS, Low TY, Lian Q, Lee CN, Mitchell W, El Oakley RM, Lim SK. Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Mol Cell Proteomics. 2007;6:1680–1689. doi: 10.1074/mcp.M600393-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gnecchi M, He H, Liang OD, Melo LG, Morello F, Mu H, Noiseux N, Zhang L, Pratt RE, Ingwall JS, Dzau VJ. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat Med. 2005;11:367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- 74.Gnecchi M, He H, Noiseux N, Liang OD, Zhang L, Morello F, Mu H, Melo LG, Pratt RE, Ingwall JS, Dzau VJ. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20:661–669. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- 75.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 76.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 77.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 79.Rosell R, Wei J, Taron M. Circulating microRNA Signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin Lung Cancer. 2009;10:8–9. doi: 10.3816/CLC.2009.n.001. [DOI] [PubMed] [Google Scholar]
- 80.Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomasoni S, Longaretti L, Rota C, Morigi M, Conti S, Gotti E, Capelli C, Introna M, Remuzzi G, Benigni A. Transfer of growth factor receptor mRNA via exosomes unravels the regenerative effect of mesenchymal stem cells. Stem Cells Dev. 2013;22:772–780. doi: 10.1089/scd.2012.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O’Brien D, van Wijnen AJ, Lerman LO. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551:55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim K, Hung RJ, Perrimon N. miR-263a regulates ENaC to maintain osmotic and intestinal stem cell homeostasis in Drosophila. Dev Cell. 2017;40:23–36. doi: 10.1016/j.devcel.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ionescu L, Byrne RN, van Haaften T, Vadivel A, Alphonse RS, Rey-Parra GJ, Weissmann G, Hall A, Eaton F, Thebaud B. Stem cell conditioned medium improves acute lung injury in mice: in vivo evidence for stem cell paracrine action. Am J Physiol Lung Cell Mol Physiol. 2012;303:L967–977. doi: 10.1152/ajplung.00144.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maron-Gutierrez T, Silva JD, Asensi KD, Bakker-Abreu I, Shan Y, Diaz BL, Goldenberg RC, Mei SH, Stewart DJ, Morales MM, Rocco PR, Dos Santos CC. Effects of mesenchymal stem cell therapy on the time course of pulmonary remodeling depend on the etiology of lung injury in mice. Crit Care Med. 2013;41:e319–333. doi: 10.1097/CCM.0b013e31828a663e. [DOI] [PubMed] [Google Scholar]
- 86.Pizurki L, Zhou Z, Glynos K, Roussos C, Papapetropoulos A. Angiopoietin-1 inhibits endothelial permeability, neutrophil adherence and IL-8 production. Br J Pharmacol. 2003;139:329–336. doi: 10.1038/sj.bjp.0705259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gamble JR, Drew J, Trezise L, Underwood A, Parsons M, Kasminkas L, Rudge J, Yancopoulos G, Vadas MA. Angiopoietin-1 is an antipermeability and anti-inflammatory agent in vitro and targets cell junctions. Circ Res. 2000;87:603–607. doi: 10.1161/01.res.87.7.603. [DOI] [PubMed] [Google Scholar]
- 88.Fang X, Neyrinck AP, Matthay MA, Lee JW. Allogeneic human mesenchymal stem cells restore epithelial protein permeability in cultured human alveolar type II cells by secretion of angiopoietin-1. J Biol Chem. 2010;285:26211–26222. doi: 10.1074/jbc.M110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JW, Fang X, Gupta N, Serikov V, Matthay MA. Allogeneic human mesenchymal stem cells for treatment of E. coli endotoxin-induced acute lung injury in the ex vivo perfused human lung. Proc Natl Acad Sci U S A. 2009;106:16357–16362. doi: 10.1073/pnas.0907996106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815–1822. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 91.Bochev I, Elmadjian G, Kyurkchiev D, Tzvetanov L, Altankova I, Tivchev P, Kyurkchiev S. Mesenchymal stem cells from human bone marrow or adipose tissue differently modulate mitogen-stimulated B-cell immunoglobulin production in vitro. Cell Biol Int. 2008;32:384–393. doi: 10.1016/j.cellbi.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 92.Corcione A, Benvenuto F, Ferretti E, Giunti D, Cappiello V, Cazzanti F, Risso M, Gualandi F, Mancardi GL, Pistoia V, Uccelli A. Human mesenchymal stem cells modulate B-cell functions. Blood. 2006;107:367–372. doi: 10.1182/blood-2005-07-2657. [DOI] [PubMed] [Google Scholar]
- 93.Ortiz LA, Dutreil M, Fattman C, Pandey AC, Torres G, Go K, Phinney DG. Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proc Natl Acad Sci U S A. 2007;104:11002–11007. doi: 10.1073/pnas.0704421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nemeth K, Leelahavanichkul A, Yuen PS, Mayer B, Parmelee A, Doi K, Robey PG, Leelahavanichkul K, Koller BH, Brown JM, Hu X, Jelinek I, Star RA, Mezey E. Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med. 2009;15:42–49. doi: 10.1038/nm.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Caser EB, Zandonade E, Pereira E, Gama AM, Barbas CS. Impact of distinct definitions of acute lung injury on its incidence and outcomes in Brazilian ICUs: prospective evaluation of 7, 133 patients. Crit Care Med. 2014;42:574–582. doi: 10.1097/01.ccm.0000435676.68435.56. [DOI] [PubMed] [Google Scholar]
- 96.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 100.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer. 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang YP, Li KB. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]