Figure 6.
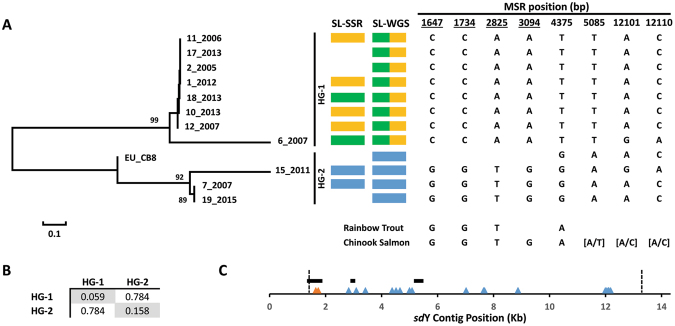
Male specific region SNP haplotypes. Male Specific Region SNP haplotypes were used to estimate evolutionary distance (substitutions per site) and construct a neighbor-joining tree linking male fish (A). Animal identifiers are shown to the right of each branch, and nodes with bootstrap replication exceeding 80% are shown to the left. The topography of the tree revealed two clear haplogroups, named HG-1 and HG-2. The assignment of males to sex lineages, using either simple sequence repeats (SL_SSR,7) or whole genome sequencing (SL-WGS, this study), is indicated by blue (SL-02), green (SL-03) or yellow (SL-06) blocks. WGS was unable to discriminate SL-03 from SL-06, and is therefore represented by both green and yellow. Alleles are shown for 8 positions where homology testing assigned the nucleotide present in either Rainbow trout or Chinook salmon. Phylogenetically informative positions relating to the ancestral state in Atlantic salmon are underlined, and those retaining polymorphism are denoted using square brackets. (B) The genetic distance present within each haplogroup is given as the mean number of substitutions per polymorphic site (grey boxes). The distance separating the haplogroups is given using the same metric. (C) A total of 22 polymorphic MSR sites were used to construct the tree. Their position within the 20 Kb sdY contig is shown using triangles. sdY exons 2–4 are shown using back triangles. The boundary of the MSR is indicated using vertical dashed lines. Two SNP (orange) overlap with sdY exon 2 while the remaining 20 are intergenic (blue).