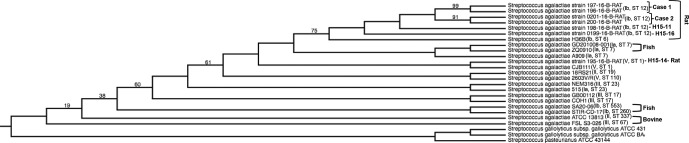
Fig. 4.
Whole-genome phylogeny comparing different species-derived GBS strains. The phylogenetic tree was generated using rat-derived GBS isolates (labelled as 'Case 1', 'Case 2', 'H15-11', H15'14 and 'H15-16') and streptococcal genomes derived from different species. Serotype and sequence type (ST) are labelled next to each individual genome. Species derivation is indicated next to rat, fish and bovine isolates. The remainder of unmarked S. agalactiae isolates are human-derived genomes. The align and tree were constructed using the pipeline Phylogenomic Estimation with Progressive Refinement (PEPR). Numbers indicate branch support values.