Abstract
A number of mutations in genes that encode ubiquitously expressed RNA-binding proteins cause tissue specific disease. Many of these diseases are neurological in nature revealing critical roles for this class of proteins in the brain. We recently identified mutations in a gene that encodes a ubiquitously expressed polyadenosine RNA-binding protein, ZC3H14 (Zinc finger CysCysCysHis domain-containing protein 14), that cause a nonsyndromic, autosomal recessive form of intellectual disability. This finding reveals the molecular basis for disease and provides evidence that ZC3H14 is essential for proper brain function. To investigate the role of ZC3H14 in the mammalian brain, we generated a mouse in which the first common exon of the ZC3H14 gene, exon 13 is removed (Zc3h14Δex13/Δex13) leading to a truncated ZC3H14 protein. We report here that, as in the patients, Zc3h14 is not essential in mice. Utilizing these Zc3h14Δex13/Δex13mice, we provide the first in vivo functional characterization of ZC3H14 as a regulator of RNA poly(A) tail length. The Zc3h14Δex13/Δex13 mice show enlarged lateral ventricles in the brain as well as impaired working memory. Proteomic analysis comparing the hippocampi of Zc3h14+/+ and Zc3h14Δex13/Δex13 mice reveals dysregulation of several pathways that are important for proper brain function and thus sheds light onto which pathways are most affected by the loss of ZC3H14. Among the proteins increased in the hippocampi of Zc3h14Δex13/Δex13 mice compared to control are key synaptic proteins including CaMK2a. This newly generated mouse serves as a tool to study the function of ZC3H14 in vivo.
Introduction
Post-transcriptional processing of mRNA, which is critical to ensure proper gene expression, is mediated by numerous RNA-binding proteins that associate with RNA from the start of transcription in the nucleus to degradation in the cytoplasm (1,2). There are a growing number of examples where mutations in genes that encode ubiquitously expressed RNA-binding proteins required for post-transcriptional processing cause tissue-specific human disease (3). Many of these mutations cause diseases of the nervous system such as fragile X syndrome (4) and spinal muscular atrophy (5). Understanding the functions of these RNA-binding proteins within the nervous system is an important step towards defining the molecular mechanism underlying these tissue-specific diseases.
Fine-tuning of the size and number of synaptic connections in the brain is critical for cognition and underlies fundamental processes such as learning and memory (6). One key mechanism by which neurons finely tune synapses is through spatio-temporal regulation of gene expression at the site of the synapse (7,8). Much of this regulation is mediated by RNA-binding proteins that modulate steps in post-transcriptional processing of mRNAs to achieve local protein synthesis in neurites (7). A number of mRNA binding proteins, such as Fragile X Mental Retardation Protein (FMRP) and Zipcode Binding Protein 1 (ZBP1), regulate local protein synthesis (8) in neurons. FMRP binds to target mRNAs and represses their local translation, allowing for finely-tuned expression of key synaptic plasticity proteins (9). ZBP1 guides target mRNAs to the growth cone and synchronizes their translation in response to external stimuli (8). Such specific functions that may be most essential in neurons could explain why mutations in genes encoding ubiquitously expressed RNA-binding proteins commonly cause neurological defects.
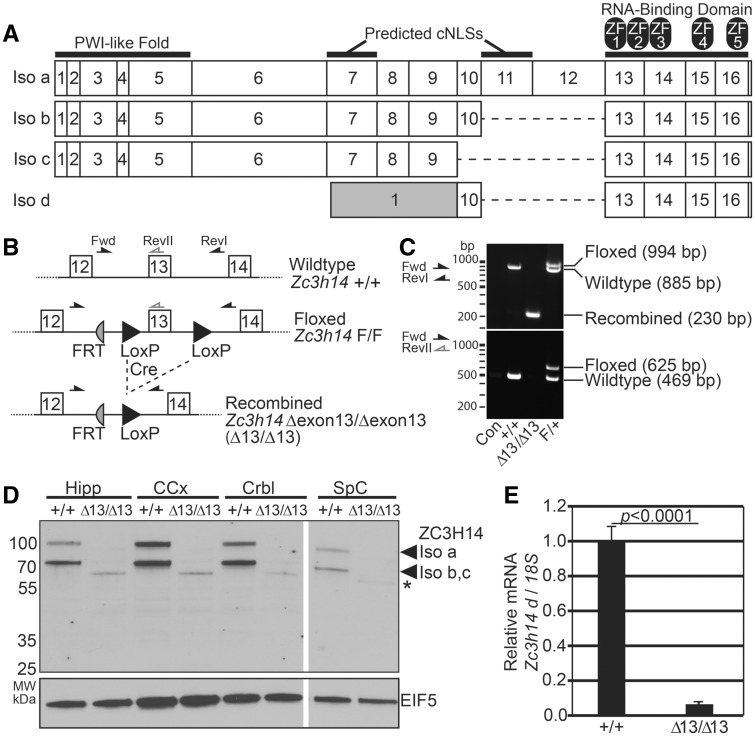
Recently, mutations in the gene encoding the evolutionarily conserved polyadenosine RNA-binding protein, ZC3H14 (Zinc finger CysCysCysHis protein 14), were identified as the cause of a form of nonsyndromic, autosomal recessive intellectual disability (10). ZC3H14 is evolutionarily conserved (10–14), and orthologues are essential in budding yeast (15) and flies (10). The critical functionally conserved domains present in ZC3H14 (Fig. 1A) include an N-terminal Proline-Tryptophan-Isoleucine (PWI)-like domain that mediates interactions with the nuclear pore (16), predicted nuclear targeting signals, and a C-terminal tandem CysCysCysHis (CCCH) Zinc Finger (ZF) domain that mediates high affinity binding to polyadenosine RNA (17). As shown in Figure 1A, alternative splicing in mammals generates several isoforms of ZC3H14 (isoforms a-d in mice) including one, isoform d, that lacks the exons that encode the N-terminal PWI domain and nuclear targeting signals.
Figure 1.
Generation of Zc3h14Δex13/Δex13 mice. (A) Schematic of four protein isoforms of murine Zc3h14 gene (Isoforms a-d). All isoforms generated contain the C-terminal RNA-binding domain, composed of five CCCH (Cys-Cys-Cys-His) zinc fingers (labelled as ZF1-5). Isoforms a-c contain the N-terminal PWI-like fold, important for poly(A) RNA export from nucleus in budding yeast (28) and predicted classical nuclear localization signal (NLS) motifs. Isoform d contains an alternative first exon (gray) that splices directly to exon 10 encoding a smaller protein isoform. Note that exon 13 is the first common exon among all Zc3h14 splice variants and encodes the beginning of the critical RNA-binding domain (13,17). (B) A map illustrating the location of the loxP and FRT sites adjacent to Zc3h14 exon 13 in the floxed allele is shown. The recombined allele illustrates the removal of Zc3h14 exon 13 following Cre-mediated recombination. The positions of the forward (Fwd) and reverse (RevI, RevII) primers used for genotyping are indicated. FRT, Flippase Recognition Target; +, control allele; F, floxed allele; Δexon13 or Δex13, recombined allele lacking exon 13. (C) Genotyping of the Zc3h14 allele by PCR using primers illustrated in (B). Representative agarose gels are shown for PCR (top gel) using the primers flanking Zc3h14 exon 13, including LoxP sites (Fwd and RevI, black arrows), or (bottom gel) using the Fwd primer and reverse primer located in exon 13 (RevII, grey reverse arrow). Results are shown for no genomic DNA control (Con); a +/+ control mouse; a Δex13/Δex13 (Δ/Δ) homozygous recombined mouse; and a heterozygous floxed/+ (F/+) mouse. The size of the products generated is indicated to the right, and markers in bp are shown on the left. (D) ZC3H14 protein was analysed in tissue samples collected from Zc3h14+/+ or Zc3h14Δex13/Δex13 mouse hippocampus (Hipp), cerebral cortex (CCx), cerebellum (Crbl), and spinal cord (SpC). The top panel shows an immunoblot for ZC3H14 using an N-terminal antibody that recognizes the PWI-like fold (14). The band corresponding to isoform a is located at ∼100 kDa; and the shorter isoforms, b and c, are detected as a single band at ∼70 kDa. The bottom panel shows an immunoblot for EIF5, eukaryotic Initiation Factor 5, as a loading control (101). (*) denotes a truncated N-terminal fragment of ZC3H14 (see Supplementary Material, Fig. S1). (E) Quantitative PCR analysis of Zc3h14 splice variant d (which lacks the PWI-like Fold recognized by the antibody), normalized to 18S rRNA as an internal control, isolated from Zc3h14+/+ or Zc3h14Δex13/Δex13mouse brain. Error bars indicate SEM. P < 0.0001.
Patients that suffer from intellectual disability caused by mutation of ZC3H14 are homozygous for a ZC3H14 allele with a premature stop codon in exon 6 (R154X) that eliminates expression of the protein isoforms that most resemble the essential orthologues found in yeast (15) and flies (10). ZC3H14 isoform d, which lacks exon 6, is unaffected by the mutation (14). Although this form of the protein is localized to the cytoplasm and appears to be expressed primarily in testes (14), possibly this short ZC3H14 isoform could fulfill an essential function in these patients (18). While there could be a functional orthologue of ZC3H14 that does not share extensive amino acid similarity in key domains, the human genome does not encode any other CCCH zinc finger proteins that contain a PWI-like domain or key conserved residues within the zinc finger domain that are critical for RNA binding in ZC3H14 orthologues (19), raising the possibility that ZC3H14 function may not be essential in mammals. Prior to the current study, there was no way to assess the requirement for mammalian ZC3H14.
ZC3H14 binds with high-affinity to polyadenosine RNA (10,13). Studies in cultured human cell lines show that ZC3H14 is localized primarily to nuclear speckles (14,20) suggesting functions within the nucleus. In fact, characterization of ZC3H14 orthologues in model organisms reveals a number of nuclear functions for this zinc finger polyadenosine RNA-binding protein including reported roles in transcription (21), control of poly(A) tail length (12,22), splicing (23), and RNA export from the nucleus (24). These functions are consistent with the steady-state localization of the protein to the nucleus and the confirmed nucleocytoplasmic shuttling of the S. cerevisiae orthologue (25–27). However, such functions alone cannot readily explain why loss of ZC3H14 in patients leads to brain-specific deficits. The specific cellular requirement for ZC3H14 has been addressed by studies exploiting loss-of-function models of the Drosophila orthologue, dNab2. Flies lacking both zygotic and maternally-deposited dNab2 are not viable (10) while zygotic mutants show reduced viability as well as defects in brain morphology (11). Neuron-specific knockdown of dNab2 impairs short-term memory as determined in a courtship conditioning assay (11). Furthermore, neuron-specific re-expression of dNab2 or expression of human ZC3H14 is sufficient to rescue behavioral defects in dNab2 zygotic mutant flies (10,12). These studies establish a critical, functionally conserved requirement for Drosophila ZC3H14 (dNab2) in neurons.
To examine the function of ZC3H14 in a mammalian brain and provide insight into why a loss-of-function mutation in ZC3H14 causes brain-specific defects in humans, we generated mice that lack Zc3h14 exon 13, an exon common to all Zc3h14 splice variants (denoted as Zc3h14Δex13/Δex13). Here, we report an evolutionarily conserved function of ZC3H14 in poly(A) tail length control in the mammalian brain. Furthermore, Zc3h14Δex13/Δex13mice show morphological changes in ventricles within the brain as well as a deficit in working memory. Quantitative label-free proteomic analysis reveals key pathways with roles in synaptic function affected upon the loss of ZC3H14. The Zc3h14Δex13/Δex13 mouse and these findings reveal the importance of ZC3H14 within the mammalian brain and provide an important tool to further define the role of ZC3H14 in the brain.
Results
Generation and confirmation of Zc3h14Δex13/Δex13 mice
As with humans (14), mice express multiple splice variants of the Zc3h14 gene, which encode four protein isoforms termed ZC3H14 isoforms a-d (Fig. 1A). These isoforms share 90% amino acid sequence identity with the human ZC3H14 isoforms. Exon 13 is the first common exon among the Zc3h14 splice variants. This exon encodes the start of the zinc finger domain that mediates binding to RNA (10,13,14,17,19,28) and is essential for the function of the budding yeast ZC3H14 orthologue, Nab2 (24). As described in Materials and Methods, we obtained embryonic stem cells in which Zc3h14 exon 13 is flanked by loxP sites from the Knockout Mouse Project (Fig. 1B). These cells were utilized to generate a conditional knockout mouse line expressing a floxed allele for Zc3h14 which was induced to create an out-of-frame deletion of Zc3h14 exon 13 (termed Δexon13 or Δex13) upon Cre-mediated recombination.
To examine the function of ZC3H14, we inactivated Zc3h14 by mating homozygous floxed Zc3h14F/F mice to EIIa-Cre transgenic mice, which express Cre-recombinase under the control of a strong adenovirus EIIa promoter in a wide range of tissues in the mouse embryo including germ cells (29,30). EIIa-Cre transgenic mice are commonly used to generate germ-line deletion of loxP-flanked genes (31–37). Following several generations of breeding as described in Materials and Methods, the floxed Zc3h14 conditional allele was efficiently recombined to yield EIIa-Zc3h14Δex13/Δex13 mice as determined by genomic PCR analysis (primers indicated in Fig. 1B) to detect the control, floxed, and recombined alleles. The recombination event was confirmed by the appearance of a lower molecular weight band at the predicted size (230 bp) upon Cre-mediated deletion of exon 13 (Fig. 1C, top panel). Additional PCR analyses confirmed the loss of exon 13 in the recombined mice (Fig. 1C, bottom panel). Mice with confirmed recombination were mated to wildtype mice to breed out the EIIa-Cre allele. EIIa-Cre-negative, Zc3h14Δex13/+ mice were mated to generate Zc3h14Δex13/Δex13 homozygous mice for a minimum of four generations for subsequent analyses.
To validate the loss of ZC3H14 protein in these mice, we immunoblotted tissue isolated from the hippocampus, cerebral cortex, cerebellum, and spinal cord, using an antibody that detects the N-terminal PWI-like domain of ZC3H14 (14). As shown in Figure 1D, the Zc3h14Δex13/Δex13 mice show no detectable expression of ZC3H14 isoforms a-c, which are readily detectable in Zc3h14+/+control mice. Note that we did detect a small amount of a lower molecular weight band that is more evident in some tissues than others (asterisk in Fig. 1D), which is specifically recognized by the N-terminal ZC3H14 antibody. To identify the protein corresponding to this band, we immunoprecipitated using ZC3H14 antibody and analysed the precipitated protein by mass spectrometry. This analysis reveals that this lower molecular weight band corresponds to a truncated form of ZC3H14. ZC3H14 peptides detected in the immunoprecipitate from the Zc3h14Δex13/Δex13 mouse map only to the N-terminal region of the ZC3H14 protein within exons 5 and 6 (Supplementary Material, Fig. S1), suggesting low-level expression of a truncated protein that lacks the essential RNA-binding domain (24). In contrast, peptides detected in the immunoprecipitate from the control mouse map to both the N- and C-terminal regions of the ZC3H14 protein (Supplementary Material, Fig. S1). Furthermore, the amount of ZC3H14 immunoprecipitated from Zc3h14+/+ mice is at least ten-fold more than the amount of truncated protein immunoprecipitated from Zc3h14Δex13/Δex13mice showing that the level of this truncated protein is very low. Thus, Zc3h14Δex13/Δex13mice do express a small amount of truncated ZC3H14, which is devoid of the functionally essential RNA-binding zinc finger domain (13,24). As no antibody is available to detect ZC3H14 isoform d by immunoblotting, we performed real-time PCR analysis of mouse brain to confirm that this variant was also absent from the Zc3h14Δex13/Δex13 mice. The Zc3h14 isoform d transcript was not detectable above background in Zc3h14Δex13/Δex13 mice, but was readily detected in Zc3h14+/+ mice (Fig. 1E). Taken together, these analyses confirm that we have generated mice lacking any full-length ZC3H14 isoform.
ZC3H14 is not essential but is required for normal litter and testis size
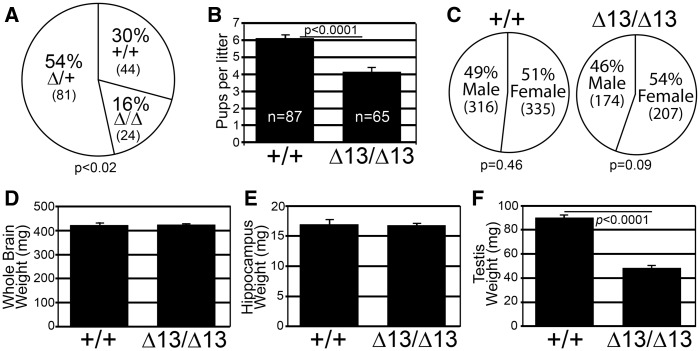
To assess the requirement for ZC3H14 in mice, we bred heterozygous Zc3h14Δex13/+mice to one another, calculated ratios of the genotypes produced, and compared them to expected Mendelian ratios. While Zc3h14Δex13/Δex13mice are viable, there was a statistically significant difference between the expected (1:2:1) and observed (1.00:1.8:0.6, P < 0.02) ratios of the genotypes produced (Fig. 2A). To assess differences in litter size, we bred homozygous Zc3h14Δex13/Δex13mice to one another. We detected a modest but statistically significant decrease in average litter size (4.1 pups/litter) generated from these Zc3h14Δex13/Δex13pairings when compared with Zc3h14+/+pairings (6.1 pups/l) (Fig. 2B). This decrease in litter size may at least partially be due to a trend toward a decrease in the number of male Zc3h14Δex13/Δex13births as shown in Figure 2C. Together, these data indicate that although Zc3h14 is not essential for viability, loss of ZC3H14 could impair survival in utero or alter maternal care.
Figure 2.
Zc3h14Δex13/Δex13 mice are viable. (A) Zc3h14 genotype percentages produced by heterozygous Zc3h14Δ/+ breeding pairs. The total numbers of mice obtained for each genotype: +/+ wildtype; Δ/+ heterozygous; and Δ/Δ homozygous Δex13 are indicated in parentheses. P = 0.0.02. (B) The average number of pups per litter generated from Zc3h14+/+ (+/+) pairings or Zc3h14Δex13/Δex13 (Δ13/Δ13) parings are indicated in the bar graph. P < 0.0001. (C) The percent of males and females of Zc3h14+/+ and Zc3h14Δex13/Δex13 mice born from Zc3h14+/+ pairings or Zc3h14Δex13/Δex13 parings is presented by genotype. Means of (D) whole brain weight and (E) hippocampus weight for Zc3h14+/+ (+/+) and Zc3h14Δex13/Δex13 (Δ13/Δ13) adult male mice are plotted. Zc3h14+/+, n = 3; Zc3h14Δex13/Δex13, n = 3. (F) Testis weight (mg) comparison between Zc3h14+/+ (+/+) and Zc3h14Δex13/Δex13 (Δ13/Δ13) adult mice are plotted. P < 0.0001. Zc3h14+/+, n = 9; Zc3h14Δex13/Δex13, n = 9. Error bars indicate SEM for (B,D–F).
As a general indication of overall health and development, we systematically measured the body weight of the mice starting at 3 weeks after birth and continuing until they were 5 months old (Supplementary Material, Fig. S2A). Analysis of mean values for body weight of male and female Zc3h14+/+ and Zc3h14Δex13/Δex13 mice showed no statistically significant difference between the two genotypes. The sample size, standard error of the means (SEMs), and P values are reported in Supplementary Material, Figure S2B–D. To assess any gross impact on the brain, we also measured both total brain weight and hippocampal weight. We did not detect any statistically significant difference between the mean values for whole brain weight (Fig. 2D) or hippocampal weight (Fig. 2E) when we compared male Zc3h14+/+ and Zc3h14Δex13/Δex13 adult mice.
As the testes express high levels of ZC3H14 (14), we compared testis weight in Zc3h14+/+ versus Zc3h14Δex13/Δex13 mice. Although no gross differences in body weight or brain weight were detected, Zc3h14Δex13/Δex13mice had testes that were half the weight of Zc3h14+/+ testes (Fig. 2F). ZC3H14 is thus required for normal litter size and is important for normal testis size.
Zc3h14Δex13/Δex13mice show altered ventricles in the brain
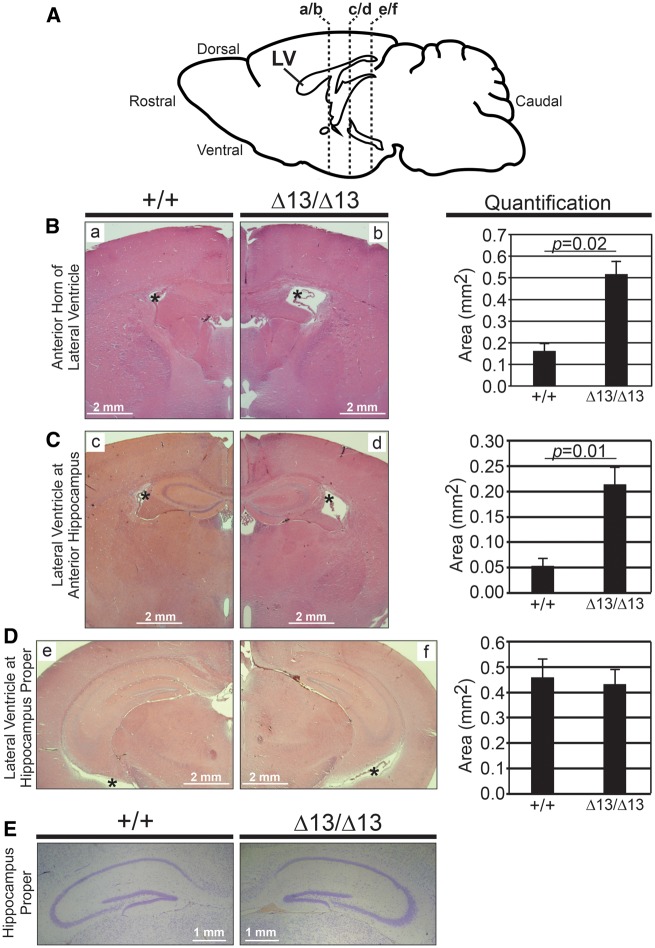
Although the brain and hippocampi of Zc3h14Δex13/Δex13 mice showed no detectable change in weight as compared to Zc3h14+/+ mice (Fig. 2D and E), we examined overall brain morphology by histology. We analysed coronal histological sections (as diagrammed in Fig. 3A) comparing Zc3h14+/+and Zc3h14Δex13/Δex13mouse brains. Haematoxylin and eosin (H&E) staining revealed that the anterior portion of the lateral ventricles is enlarged in Zc3h14Δex13/Δex13mice as compared to Zc3h14+/+ mice (Fig. 3B and C). In contrast, there was no detectable effect on the size of the lateral ventricles at a more caudal level at the hippocampus proper (Fig. 3D). To examine hippocampal morphology, we stained matched sections from Zc3h14+/+and Zc3h14Δex13/Δex13 mice with cresyl violet (Fig. 3E). At this level of analysis, no gross morphological differences were detected between the Zc3h14+/+and Zc3h14Δex13/Δex13 hippocampi when sections from five independent mice were examined.
Figure 3.
Analysis of brain and hippocampal morphology in Zc3h14Δex13/Δex13 mice. (A) Diagram of lateral view of mouse brain and location of coronal sections shown in the following histological images. LV, lateral ventricle. Representative H&E stains of lateral ventricles of Zc3h14+/+ (+/+) and Zc3h14Δex13/Δex13 (Δ13/Δ13) adult mice at the following level of slices: (B) anterior horns of ventricles (a/b in diagram), (C) anterior hippocampus (c/d in diagram), and (D) hippocampus proper (e/f in diagram). Magnification ×2. Scale bars, 2 mm. *, lateral ventricle. Shown to the right are the corresponding quantifications of the area (mm2) of the lateral ventricles at the indicated levels for Zc3h14+/+ and Zc3h14Δex13/Δex13mice. Error bars indicate SEM. For quantification of anterior horns of lateral ventricles; Zc3h14+/+, n = 3; Zc3h14Δex13/Δex13, n = 5. For quantification of lateral ventricles at anterior hippocampus; Zc3h14+/+, n = 3; Zc3h14Δex13/Δex13, n = 3. For quantification of lateral ventricles at hippocampus proper; Zc3h14+/+, n = 6; Zc3h14Δex13/Δex13, n = 6. (E) Comparable brain sections from Zc3h14+/+ (+/+) and Zc3h14Δex13/Δex13 (Δ13/Δ13) adult mice were stained with cresyl violet. Scale bars equal 1 mm.
Zc3h14Δex13/Δex13mice have impaired working memory but intact learning
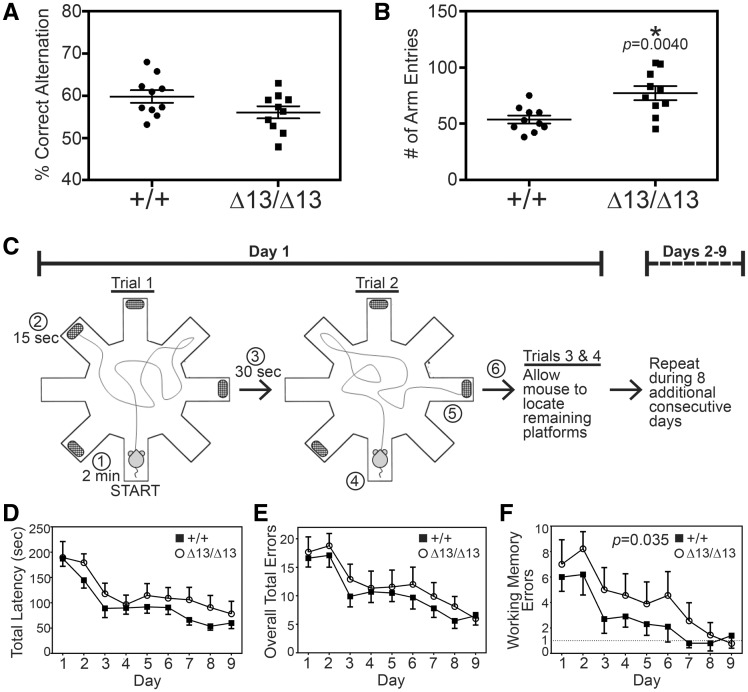
Loss of the ZC3H14 orthologue in Drosophila causes defects in short-term memory while learning remains intact (11). To assess the consequences of loss of ZC3H14 in mice, we performed a variety of behavioral assays including spontaneous alternation Y-maze, open field test, light-dark box, fear conditioning, novel cage activity, and water radial arm maze (Fig. 4 and Supplemental Material, Table S1). For all studies, we employed adult mice between 3 and 4 months of age. Independent cohorts of mice were carefully age and gender matched for all experiments. In several of the experimental paradigms including the novel cage, light-dark box, and fear conditioning, we did not detect statistically significant differences in the behaviors we examined when comparing the Zc3h14+/+ and Zc3h14Δex13/Δex13 mice (Supplementary Material, Table S1). To extend this analysis, a Y-maze paradigm was employed as an initial approach to assess working memory (38). We did not detect a statistically significant difference in the percentage of correct alternations in arm entries when we compared Zc3h14+/+ and Zc3h14Δex13/Δex13mice (Fig. 4A); however, there was a trend toward a deficit in the sequence of arm entries for Zc3h14Δex13/Δex13mice. As shown in Figure 4B, we did detect a statistically significant difference (P = 0.004) in the number of arm entries between the two groups. The Zc3h14Δex13/Δex13 mice had 40% more arm entries than the Zc3h14+/+ mice (Fig. 4B). These data suggest there are some differences in the behavior of Zc3h14Δex13/Δex13 mice compared to control Zc3h14+/+ mice.
Figure 4.
Zc3h14Δex13/Δex13 mice have impaired working memory but intact learning. Mice were first tested in a Y-maze paradigm to analyse (A) % Correct Alternation between arms and (B) number of Arm Entries. We compared Zc3h14+/+ (+/+) and Zc3h14Δex13/Δex13 (Δ13/Δ13) adult mice in 20 independent trials as described in Materials and Methods. (A) There was no statistically significant difference detected in the % Correct Alternation between arms of the maze. (B) There was a statistically significant increase (P = 0.004) in the number of arm entries (∼40% increase) for Zc3h14Δex13/Δex13 (Δ13/Δ13) compared to Zc3h14+/+ (+/+) mice. (C) Schematic of the WRAM apparatus and 9-day testing procedure (41). Mice are given 2 min to locate a hidden platform positioned at the far ends of four of eight arms. If a mouse is unable to locate a platform within 2 min of beginning the trial, it is gently guided to the nearest platform. Upon successful escape from water onto platform, mice are rewarded with 15 s of rest on platform, followed by 30 s of rest in a heated, dry cage. At the start of trial 2, the previously located platform is taken away and mice proceed to find one of the remaining hidden platforms. This process is repeated until all platforms are found, one in each trial. These four trials are repeated daily for a total of 9 consecutive days. Circled numbers indicate sequence of events in trials. The experiment compares Zc3h14+/+ (+/+, closed squares) to Zc3h14Δex13/Δex13 ((Δ13/Δ13, open circles) mice. (D) Total latency is the average time (seconds) required per subject to complete all four trials for a specified day of testing. Comparing by test day, P < 0.0001; comparing by genotype, P = 0.11, indicating no difference. (E) Overall total errors is the average number of errors made per subject for all four trials for a specified day. Comparing by test day, P < 0.0001; comparing by genotype, P = 0.20, indicating no difference. (F) The average number of working memory errors, represented by reentry into any maze arm during a given trial, per subject for a specified day, is shown. Dashed line at 1 working memory error represents sufficient performance on WRAM. Comparing by test day, P < 0.0001; comparing by genotype, P = 0.035, revealing a statistically significant difference. Open circles indicate Zc3h14Δ/Δ. Closed squares indicate Zc3h14+/+. Behavioral tests were performed on independent cohorts of 3-4-month-old mice. All tested mice were male. Zc3h14+/+, n = 10; Zc3h14Δex13/Δex13, n = 9. Error bars indicate SEM for (B), (C), (D).
Based on the results obtained in the Y-maze, we employed a more sensitive assay to further explore aspects of learning and memory, the water radial arm maze (WRAM) (39–42). As illustrated in Figure 4C, the WRAM is an eight-arm arena in which mice are evaluated on their ability to learn the location of submerged platforms in order to escape from having to swim, and remember the location of the hidden platforms. Learning is indicated, across nine days of testing, by a decrease in the amount of time required to find all four platforms and a decrease in the number of errors made. Errors include mistakes in which a mouse enters an arm that: (a) does not contain a platform, (b) contains a platform but the mouse does not locate it, (c) contained a platform on a previous trial on the same day, or (d) has already been entered previously during the trial (i.e. deficit in working memory). Both Zc3h14+/+ and Zc3h14Δex13/Δex13adult mice were able to learn the locations of the hidden platforms as evidenced by a decrease in the time required to locate the platforms (Fig. 4D, P < 0.0001), a decrease in the number of overall total errors (Fig. 4E, P < 0.0001), and a decrease in the number of working memory errors (Fig. 4F, P < 0.0001) across the course of the nine-day testing period.
Though both Zc3h14+/+ and Zc3h14Δex13/Δex13 mice show the ability to learn as assessed in the WRAM (Fig. 4D–F), data uniformly indicate a trend toward an impairment in efficient learning by the Zc3h14Δex13/Δex13mice, which require one to three additional days to perform at a similar level to Zc3h14+/+ mice. Furthermore, analysing the WRAM data to assess working memory function, Zc3h14Δex13/Δex13 mice showed a statistically significant deficit in working memory when compared with Zc3h14+/+ mice (Fig. 4F). Specifically, working memory errors are repeated entries into any arm on a given trial, indicating the inability to remember which arm was explored within the previous 2 min (the length of a single trial). As illustrated in Figure 4F, Zc3h14Δex13/Δex13mice exhibited a statistically significant (P = 0.035) persistence in the number of working memory errors that is not exhibited by control mice. Zc3h14Δex13/Δex13mice require two additional days of trials to perform as well as the control mice in locating all platforms. The WRAM results indicate that ZC3H14 is required in adult mice for proper cognitive function, particularly spatio-temporal working memory, which is consistent with the cognitive deficits seen in patients (10) and the behavioral defects observed in the dNab2 mutant flies (11).
Zc3h14Δex13/Δex13mice have normal visual function and exhibit normal motor function and coordination
The WRAM test requires vision to locate environmental cues and motor coordination to swim efficiently. To rule out possible confounding variables that could lead to impaired performance of Zc3h14Δex13/Δex13mice in the WRAM, we conducted several tests to examine visual function (Supplementary Material, Fig. S3A–C), and motor function and coordination (Supplementary Material, Fig. S3D–G). These tests were performed on independent cohorts of mice so they would not confound the results of the WRAM. Zc3h14Δex13/Δex13 mice performed at least as well as the Zc3h14+/+ mice on the visual and motor coordination assays. We measured visual acuity and contrast sensitivity by using an optokinetic test apparatus (depicted in Supplementary Material, Fig. S3A) for rodents, which elicits a head- and body-turning response to a rotating visual field (43). Zc3h14Δex13/Δex13mice showed no obvious deficit in their visual acuity represented by their ability to distinguish similar spatial frequencies of black and white gratings when compared to control mice (Supplementary Material, Fig. S3B). Furthermore, Zc3h14Δex13/Δex13 mice showed no evidence of impaired contrast sensitivity, as measured by their ability to distinguish between gradients of dark- and light-gray gratings (Supplementary Material, Fig. S3C), when compared to Zc3h14+/+ mice.
Next, we challenged mice using two assays to assess general motor function and coordination. Zc3h14Δex13/Δex13mice did not show a statistically significant difference in their ability to grip a wire mesh while up-side down (depicted in Supplementary Material, Fig. S3D) when compared to Zc3h14+/+ mice (Supplementary Material, Fig. S3E). Additionally, when challenged on a rotarod apparatus (depicted in Supplementary Material, Fig. S3F), Zc3h14Δex13/Δex13mice performed at least as well as the Zc3h14+/+ mice (Supplementary Material, Fig. S3G). These data together indicate that, along with vision, general motor function, sensorimotor coordination, and motor learning are intact in Zc3h14Δex13/Δex13mice, and thus would not contribute to differences in performance in the WRAM. Furthermore, consistent with normal baseline behavior, Zc3h14+/+ and Zc3h14Δex13/Δex13mice were comparable in assays to assess exploratory behavior (novel cage), fear conditioning, and anxiety (light/dark box) (Supplementary Material, Table S1). Together, these results indicate that the working memory deficit detected in the Zc3h14Δex13/Δex13mice is not likely to be confounded by these variables.
Zc3h14 is required for proper poly(A) tail length control
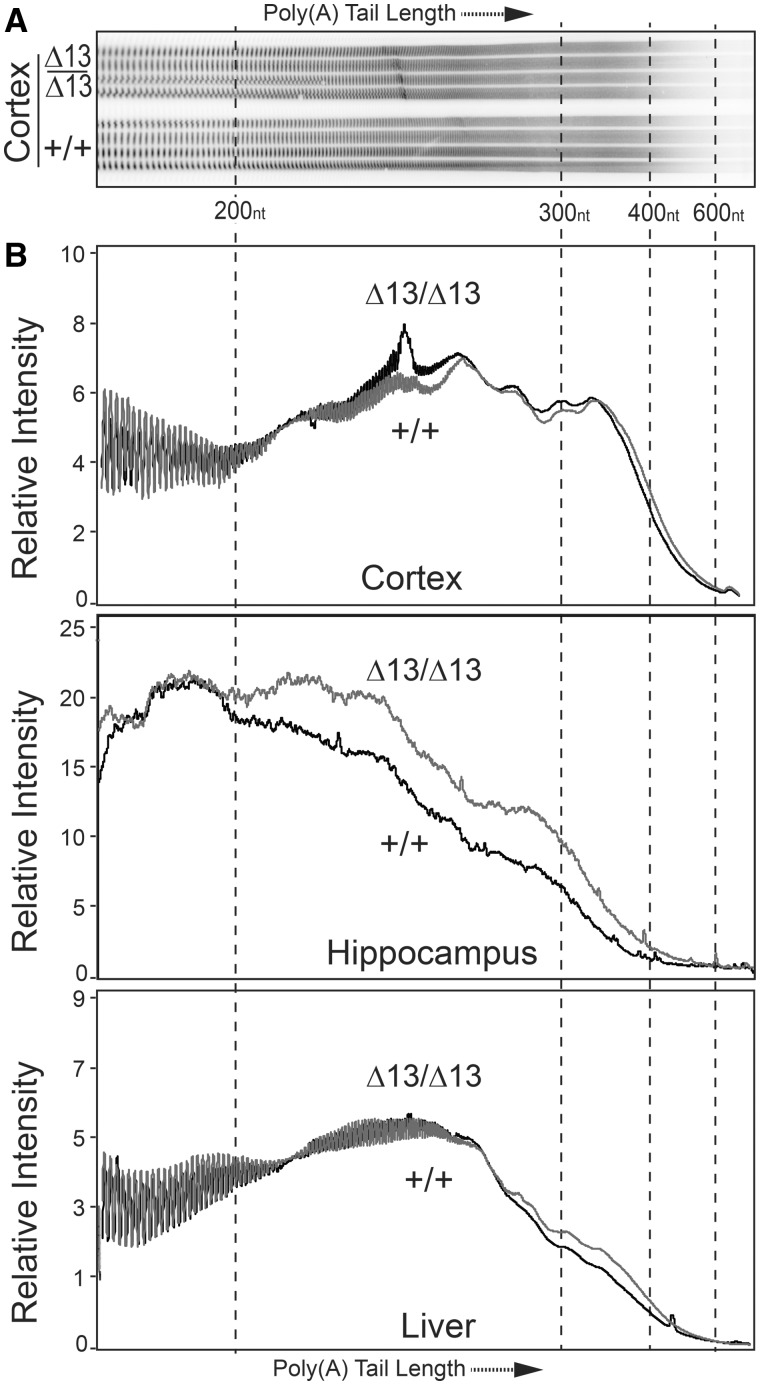
Our previous work demonstrates that one evolutionarily conserved molecular function of ZC3H14 is to restrict poly(A) tail length of bulk RNAs (10,12,15,22). However, no studies have been previously performed to assess the molecular function of vertebrate ZC3H14 in vivo. To assess the functional consequence of loss of ZC3H14, we compared poly(A) tail length of bulk RNA isolated from several different tissues of Zc3h14+/+ and Zc3h14Δex13/Δex13 mice (Fig. 5). We examined two different brain regions, cortex and hippocampus, as well as liver. ZC3H14 is highly expressed in all these tissues (14). Poly(A) tail length of bulk RNA isolated from these tissues was assessed as described in Materials and Methods using an acrylamide gel to resolve end-labeled poly(A) tracts. A typical gel is shown in Figure 5A and results depicted as scans of the lanes in each gel are shown in Figure 5B. The Zc3h14Δex13/Δex13mice show an increase in bulk poly(A) tail length compared to Zc3h14+/+ mice in all tissues examined but the effect is not statistically significant in the cortex (P = 0.11) or liver (P = 0.05). The increase in bulk poly(A) tail length is far more striking in the samples isolated from the hippocampus and reaches statistical significance based on a two-tailed Mann Whitney test (P < 0.001). This result provides molecular and in vivo evidence that ZC3H14 plays a role in poly(A) tail length control in the hippocampus.
Figure 5.
ZC3H14 is required for proper poly(A) tail length control in mice. Bulk poly(A) tail length was analysed from brain (cortex and hippocampus) and liver tissue. (A) A representative gel of the resolved bulk poly(A) sample isolated from cortex is shown for Zc3h14Δex13/Δex13 (Δ13/Δ13) and Zc3h14+/+ (+/+) hippocampal samples isolated from three independent mice. Approximate positions of nucleotide size markers (nt) are indicated. (B) To provide a quantitative measure of poly(A) tail length across samples, the Relative Intensity of the signal is plotted along the length of the poly(A) tail as determined by the size markers. The positions of the scan corresponding to 200, 300, 400, and 600 nucleotides (nt) are indicated. Results are presented for Cortex (P = 0.11), Hippocampus (P < 0.0001) and Liver (P = 0.05).
Increased expression of synaptic proteins in Zc3h14Δex13/Δex13mice
Patients with loss of function alleles of ZC3H14 have intellectual disability (10) and our mice show changes in the lateral ventricle of the brain and poly(A) tail length control in the hippocampus. To begin to explore mechanisms that could underlie these functional changes, we performed a quantitative proteomic analysis to compare the hippocampal proteomes of Zc3h14+/+and Zc3h14Δex13/Δex13mice (n = 4 each group). We focused on hippocampus for this analysis as this is the tissue that shows the most profound change in bulk poly(A) tail length (Fig. 5B). Following tissue homogenization and trypsin digestion, peptides from each sample were analysed by LC-MS/MS on an Orbitrap Fusion mass spectrometer. Relative protein abundance was determined by peptide ion-intensity measurements across LC-MS runs using the label-free quantification (LFQ) algorithm in the MaxQuant computational platform (44). In total, 56,423 peptides mapping to 4,450 protein groups were identified in Zc3h14+/+ and Zc3h14Δex13/Δex13 mouse hippocampi. The stochastic nature of LC-MS/MS based peptide sequencing required us to limit the number of missing protein measurements to no more than two per condition, resulting in the final quantification of 4,161 protein groups (henceforth referred to as proteins) mapping to 4,052 unique gene symbols (Supplemental Material, Table S2).
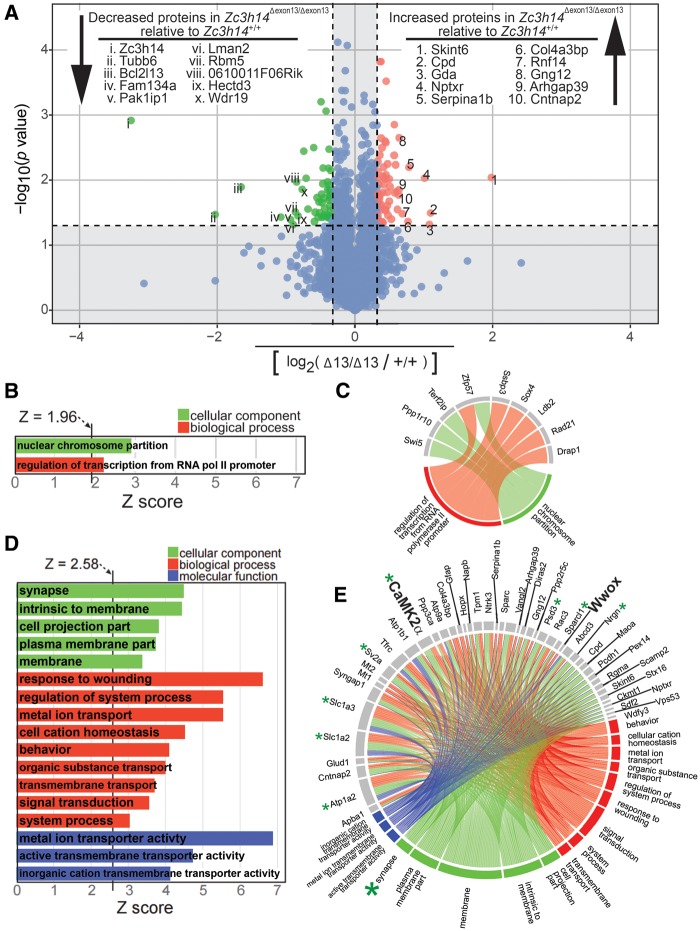
Figure 6A shows a volcano plot where each protein that has any change in steady state expression level between Zc3h14+/+and Zc3h14Δex13/Δex13hippocampi is represented by a colored dot. In total, 113 (62 increased, 51 decreased) proteins with at least a ±1.25-fold change (i.e. |log2(Zc3h14+/+/Zc3h14Δex13/Δex13)| ≥ 0.32) and a P value < 0.05 are highlighted as green (decreased) or red (increased) dots. Proteins that did not meet these criteria (shown as blue dots) fall within the shaded nonsignificant boundaries of the plot and were omitted from further analysis. The identities of the 10 most decreased and 10 most increased proteins are listed, and their corresponding positions are labeled (with Roman or Arabic numerals, respectively) on the plot. Note that the top protein with decreased expression is ZC3H14, which further validates the absence of the ZC3H14 protein from Zc3h14Δex13/Δex13 mice and highlights the reliability of our proteomics approach. We further analysed the proteins that met our significance criteria by creating a heatmap with hierarchical clustering of both samples and proteins (Supplemental Material, Fig. S4). This analysis shows that the Zc3h14+/+ and Zc3h14Δex13/Δex13 samples segregate based on protein levels, thus confirming by an independent method the distinctive proteomic profiles.
Figure 6.
Proteomic analysis of Zc3h14+/+ versus Zc3h14Δex13/Δex13 hippocampi. (A) A volcano plot of all proteins with changed expression levels between Zc3h14+/+and Zc3h14Δex13/Δex13 hippocampi, represented as colored dots is shown. Statistically significantly changed proteins are highlighted as green dots and red dots. Among these, 51 proteins (including ZC3H14) have decreased expression (green dots) in Zc3h14Δex13/Δex13 hippocampi when compared to Zc3h14+/+ hippocampi, and 63 proteins have increased expression (red dots). Inclusion criteria for statistically significance are 1.25-fold change and P < 0.05. The top 10 hits for decreased and increased proteins are listed, along with their relative positions (Roman or Arabic numerals, respectively) on the volcano plot. The remaining proteins that did not meet statistical significance are represented as blue dots and fall within the shaded region of the plot, the boundaries of which are set by the aforementioned inclusion criteria. (B) For the decreased gene set, the two GO terms along with their Z-scores are listed. Inclusion criteria for this analysis are Z-score ≥ 1.96, P value < 0.05, and ≥ 3 genes per GO term. (C) The nine decreased genes that cluster and their corresponding GO terms are shown by a connecting line. (D) For the increased gene set, the list of GO terms along with their Z-scores are listed. Inclusion criteria for this analysis are Z-score ≥ 2.58, P value < 0.01, and ≥ 5 genes per GO term. (E) The increased genes that cluster into the listed GO terms in (D) are depicted. The corresponding GO terms are shown by a connecting line. For example, the large green asterisk indicates a GO term (synapse) and small green asterisks indicates the proteins (Atp1a2, CaMK2α, Nrgn, Psd3, Slc1a2, Slc1a3, Sparcl1, Sv2a) that cluster to the GO term, also shown by connecting lines.
To analyse enrichment of proteins that participate in the same cellular components, biological processes, or molecular functions, we performed a gene ontology (GO) analysis of the decreased (50 proteins when ZC3H14 is omitted from the list) and increased (62 proteins) sets of only those proteins significantly changed in Zc3h14Δex13/Δex13 mice compared to Zc3h14+/+mice. The criteria used to cluster proteins into a GO term were Z-score ≥ 1.96, P value < 0.05, and ≥ 3 gene symbols per GO term. As shown in Figure 6B, only two GO terms were identified using the decreased protein set (n = 50), as most of these proteins do not cluster into GO terms. These GO terms are nuclear-specific and suggest these pathways are most affected in the nucleus at the protein level as a result of loss of ZC3H14. The small number of proteins that do cluster into common GO terms are depicted in Figure 6C. Thus, the majority of proteins with decreased expression do not cluster into common pathways recognized by GO terms.
We then analysed the set of increased proteins (n = 62) for GO enrichment. In striking contrast to the decreased protein set, many of the increased proteins cluster into several biologically meaningful GO terms (Fig. 6D). In fact, this result holds true even after more stringent inclusion criteria (Z-score ≥ 2.58, P value < 0.01, and ≥ 5 gene symbols per GO term) than was used for analysing the decreased protein set were applied. Many of the GO terms, such as ‘synapse,’ ‘signal transduction,’ and ‘behavior,’ have neuronal or brain-specific functions. For example, the top GO term categorized under cellular component is ‘synapse’ (large green asterisk in Fig. 6E), which contains the following gene symbols that increase in expression in Zc3h14Δex13/Δex13hippocampus when compared to Zc3h14+/+hippocampus: Atp1a2, CaMK2α, Nrgn, Psd3, Slc1a2, Slc1a3, Sparcl1, Sv2a (small green asterisks). Furthermore, the majority of the top 10 increased proteins cluster into the GO terms ‘signal transduction’ and/or ‘membrane.’ In contrast, none of the top 10 decreased proteins cluster into common GO terms. This global GO term analysis of the increased protein set suggests that mutation of Zc3h14 causes increased steady state levels of proteins in cellular components or processes important for brain function such as the synapse and signal transduction.
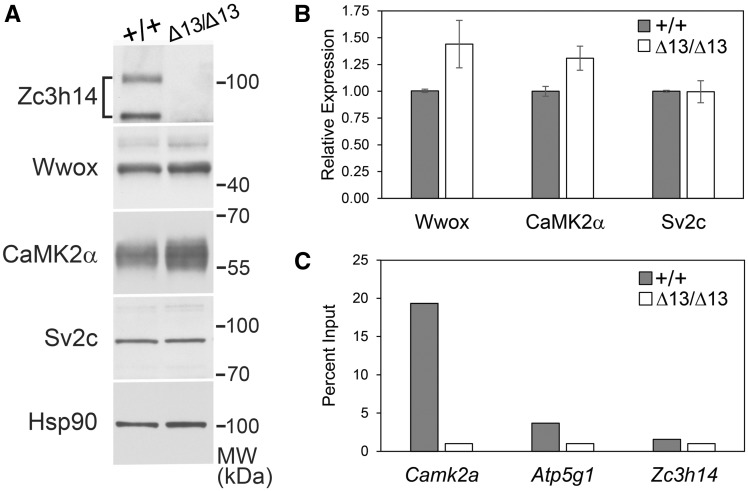
To validate the proteomic results, we selected several candidate proteins that showed a statistically significant change in the mass spectrometry data and compared expression in hippocampal lysates prepared from Zc3h14Δex13/Δex13mice to hippocampal lysates prepared from Zc3h14+/+mice (Fig. 7). We validated the decrease in the level of the ZC3H14 protein and also examined two proteins that were increase in Zc3h14Δex13/Δex13mice compared to Zc3h14+/+mice, Calcium/Calmodulin Dependent Protein Kinase II Alpha (CaMK2α/CaMKIIα) and WW domain-containing oxidoreductase (Wwox). The CaMK2α protein plays a key role in signaling within the hippocampus including a role in learning and memory (45). While most extensively characterized as a tumor suppressor, the Wwox protein has been implicated in neural development and in the pathogenesis of Alzheimer's disease (46). As a control, we also examined the expression of the Synaptic Vesicle Glycoprotein 2c (Sv2c), which showed no statistically significant difference between the mice. Results of a representative immunoblot are shown in Figure 7A. We quantitated the results of the immunoblotting using hippocampal lysates prepared from three different Zc3h14+/+or Zc3h14Δex13/Δex13mice (Fig. 7B). As predicted by the proteomic analysis, both Wwox and CaMK2α increase in Zc3h14Δex13/Δex13hippocampal samples compared to Zc3h14+/+with no detectable change in Sv2c.
Figure 7.
Validation of proteomic changes in Zc3h14+/+ compared to Zc3h14Δex13/Δex13 hippocampi. Total lysate was prepared from hippocampi of three Zc3h14+/+(+/+) and three Zc3h14Δex13/Δex13 (Δ13/Δ13) hippocampi. Samples were probed with antibodies to ZC3H14, Wwox, Cank2a, Sv2c, and a control protein Hsp90. A representative immunoblot is shown in (A) and the results from three independent samples are quantitated as a fold increase in (B). (C) To test whether ZC3H14 can bind to the CaMK2a transcript, we performed RNA immunoprecipitation from hippocampal lysates. Results are presented as fold enrichment compared to input for CaMK2a, a known target Atp5g1, and a control transcript Zc3h14.
To provide an initial test of whether proteins that show altered expression in Zc3h14Δex13/Δex13mice compared to Zc3h14+/+mice could be directly regulated by ZC3H14 at the transcript level, we performed an RNA immunoprecipitation experiment (Fig. 7C). For this experiment, we immunoprecipitated endogenous ZC3H14 from mouse hippocampi and used qRT-PCR to detect bound transcripts. We focused on CaMK2a for these initial experiments both because it is an important regulator of neuronal activity (45) and our studies in Drosophila reveal that fly ZC3H14/dNab2 can regulate the expression of a CaMK2a translational reporter and bind to the CaMK2a transcript in a comparable RNA immunoprecipitation assay (47). As shown in Figure 7C, the CaMK2a transcript is highly enriched (>15-fold) when ZC3H14 is immunoprecipitated from Zc3h14+/+hippocampus as compared to Zc3h14Δex13/Δex13hippocampus, which lacks ZC3H14. A previously validated target of ZC3H14 in cultured cells, Atp5g1 (48), is also enriched in the ZC3H14 immunoprecipitation while the Zc3h14 transcript is not. These results suggest that ZC3H14 could modulate the steady-state levels of hippocampal proteins through directly binding and regulating the corresponding transcripts.
Discussion
We have generated a Zc3h14Δex13/Δex13 mouse to study the function of the evolutionarily conserved, ubiquitously expressed polyadenosine RNA-binding protein, ZC3H14. The mutation eliminates exon 13, the first common exon of all four isoforms of ZC3H14, which encodes the start of the critical zinc finger RNA-binding domain essential for the function of the S. cerevisiae orthologue of ZC3H14, Nab2 (24). We have performed an initial characterization of the homozygous Zc3h14Δex13/Δex13 mice. Compared to control Zc3h14+/+mice, homozygous Zc3h14Δex13/Δex13mice show an increase in the size of the lateral brain ventricles with no gross change in hippocampal morphology, and modest deficits in some behavioral paradigms including the WRAM. Interestingly, bulk RNA isolated from hippocampi of Zc3h14Δex13/Δex13mice has extended poly(A) tails, and parallel quantitative proteomic analysis comparing the hippocampi of Zc3h14Δex13/Δex13mice to Zc3h14+/+mice revealed an increase in the steady-state level of proteins important for synaptic plasticity and function including CaMK2α. Taken together, this work provides a model in which to study the function of ZC3H14 in the mammalian brain.
ZC3H14 orthologues in S. cerevisiae (15) and Drosophila (10) are essential for viability. However, patients who lack expression of ZC3H14 isoforms (isoforms 1–3), which most closely resemble the evolutionarily conserved protein present in yeast and flies (18), are alive (10). Furthermore, these patients suffer from nonsyndromic intellectual disability showing that, despite ubiquitous expression of ZC3H14 isoforms 1–3, the functional consequence of loss of these protein isoforms is limited to the brain (14). Unlike the patients which retain at least one ZC3H14 isoform, the mice were engineered to remove an exon common to all splice variants and thus eliminate all protein isoforms. Although no full length ZC3H14 protein is present in these mice, we detect a very small amount of a truncated protein (Fig. 1D and Supplementary Material, Fig. S1). These results suggest that ZC3H14 is not essential for viability with the caveat that the small amount of ZC3H14 that lacks the C-terminal RNA binding domain could confer some as yet undetermined function. The Zc3h14Δex13/Δex13mice are fertile and appear to show normal growth as evidenced by analysis of overall body weight as well as brain and hippocampal weights. We did detect a decrease in litter size at weaning, which could reveal developmental consequences of loss of ZC3H14 or altered maternal care. Adult Zc3h14Δex13/Δex13 mice exhibited normal visual acuity and contrast sensitivity as well as general motor function and coordination. Taken together, these results suggest that ZC3H14 function is not critical for viability in mammals.
Two tissues that are highly dependent on spatial and temporal control of gene expression are the brain (5,49,50) and the testes (51,52). Consequently, changes in testis size are commonly associated with disorders of intellectual disability (53–57). A striking phenotype we observe in the Zc3h14Δex13/Δex13 mice is the decreased size of the testes, which suggests a requirement for ZC3H14 in proper testes development or maintenance. In fact, the testes have high expression of all ZC3H14 isoforms including the testes-enriched isoform d (14). The testes themselves are rich in RNA-binding proteins (51,52), many of which are expressed as testes-specific isoforms (58,59). Furthermore, spermatogenesis is highly dependent on post-transcriptional processing (60). Additional studies are necessary to understand the requirement for ZC3H14 in the testes and the role ZC3H14, in particular the mammalian-specific isoform d (termed isoform 4 in humans), plays in the testes.
Previous studies of the Drosophila orthologue of ZC3H14, dNab2, identified a requirement for dNab2 in supporting normal brain function and structure (11). In dNab2 mutant flies, the lobes of the mushroom bodies, neuropil which are essential for learning in flies (61), are malformed and show defects with axons inappropriately crossing the midline (11). Interestingly, such defects have also been reported in the fragile X syndrome fly model (62). Our analysis of the brain morphology of Zc3h14Δex13/Δex13 mice thus far reveals an increase in the size of the lateral ventricles with no gross change in the hippocampus. Enlargement of lateral ventricles in mouse models has been observed in a variety of brain disorders with cognitive impairment, including fragile X syndrome (63), Down syndrome (64), and the autism spectrum disorder, Smith-Lemli-Opitz syndrome (65). Furthermore, enlargement of lateral ventricles is a hallmark of developmental disability (66) and age-related cognitive decline (67,68) in humans. Many potential mechanisms could underlie the increase in the size of the lateral ventricles observed in Zc3h14Δex13/Δex13 mice including cerebrospinal fluid (CSF) overproduction, decreased resorption, or obstruction of outflow (64). Alternatively, enlarged ventricles could arise from degeneration, impaired proliferation, or delayed maturation of ependymal cells that line the ventricles (64). More detailed analyses will be required to define the nature of this defect in mice lacking ZC3H14.
With respect to the key function of ZC3H14, previous studies established that ZC3H14 and its orthologues play an evolutionarily conserved role in regulating poly(A) tail length of RNAs in budding yeast (22), Drosophila (10), and a murine neuroblastoma cell line (12). Utilizing the Zc3h14Δex13/Δex13 mice, we present the first functional characterization of the mammalian ZC3H14 protein in vivo. Our results reveal that ZC3H14 is required for proper control of poly(A) tail length in the hippocampus. Control of poly(A) tail length is crucial for proper post-transcriptional regulation (69), especially in neurons that require precise temporal and spatial control of gene expression (70–74). An elongated poly(A) tail could increase the half-life (69) of specific target RNAs and/or increase translation through a cytoplasmic polyadenylation element binding protein (CPEB)-dependent mechanism (75,76). Either mechanism could lead to an increase steady state level of the encoded protein. In fact, GO-term analysis of our mass spectrometry results revealed a unilateral increase in the expression of proteins (e.g. CaMK2α) important for processes such as signal transduction and synaptic function. Such changes could be mediated by direct binding of ZC3H14 to target transcripts as ZC3H14 enriches the CaMK2a transcript in RNA immunoprecipitation.
Results of the behavioral analyses reveal that the Zc3h14Δex13/Δex13 mice apparently have intact spatio-temporal learning but a detectable deficit in working memory revealed in the WRAM. These data are remarkably consistent with studies in a Drosophila model, in which flies depleted of dNab2 specifically in neurons, show defects in short-term memory but preserved learning ability (11). Working memory is closely related to attention (77), and measures of working memory capacity predict higher performance on executive function tasks (78). Not only is working memory critical to brain development (79), but it is also a marker for age-related decline in cognitive function (80). Taken together, it is possible that the human phenotype is, at least in part, recapitulated by the behavioral results in our rodent model.
This initial characterization of Zc3h14Δex13/Δex13 mice reveal some parallel results to mice lacking the RNA binding protein Fmr1, which serve as a model of fragile X syndrome (81). Studies in Drosophila support a close functional link as well as genetic interactions between dNab2 and dFmr1 (47). Many years of analysis of the Fmr1 knockout mice have identified changes at both the molecular and behavioral levels (81) some of which can be compared to this initial characterization of Zc3h14Δex13/Δex13 mice. While Fmr1 knockout mice have not been analysed in the WRAM, some studies report deficits in either the Morris water maze or in maze learning while others found no difference (81). Studies also revealed no statistically significant difference in a novel object recognition paradigm for Fmr1 knockout mice compared to control (81) as we report here for Zc3h14Δex13/Δex13 mice. The extensive behavioral studies performed with Fmr1 mice provide direction for future analyses of the mice lacking ZC3H14.
Another point of comparison for the Zc3h14Δex13/Δex13 mice and the Fmr1 knockout mice is proteomic changes that occur in the absence of these key regulatory RNA binding proteins. While an analysis of the hippocampal proteome directly comparable to the one reported here for Zc3h14Δex13/Δex13 has not been carried out for Fmr1 knockout mice, there are proteomic studies that analysed primary cortical neurons (82) or synaptic membranes (83,84). The only study that employed samples derived from the hippocampus used iTRAQ analysis (83) and detected low fold changes with the largest difference 1.24-fold. That study identified 23 proteins that met the criteria set forth to define statistically significant changes. We detected all 23 of these proteins in our analysis of the Zc3h14Δex13/Δex13 hippocampal proteome, and of these 23, we detected changes in 18 with the overlap primarily among proteins that show an increase in steady state levels in the mutant compared to the wildtype control animals. Some of these changes did not meet our strict criteria for statistical significance so are not reported in the Supplemental Data. While the previous study detected an increase in CaMK2α levels in the Fmr1 proteomic analysis, they were unable to validate a statistically significant change by immunoblotting. However, other studies have identified CaMK2a as an RNA regulated by FMRP/Fmr1 in multiple experimental systems (47,85). These results are consistent with our finding that CaMK2α protein levels increase in the hippocampus of Zc3h14Δex13/Δex13 mice compared to control and that CaMK2a mRNA is enriched in an RNA immunoprecipitation of ZC3H14 from hippocampus. These results support the possibility that ZC3H14 could regulate some of the same target RNAs as FMRP that are critical for proper neuronal function.
In conclusion, we have generated a Zc3h14Δex13/Δex13 mouse as a tool to study the role ZC3H14 plays in the mammalian brain. We establish here that ZC3H14 is required for proper brain function in mice and also demonstrate that ZC3H14 plays a conserved role in poly(A) tail length control within the hippocampus. Based on our proteomic analysis, we suggest a model where ZC3H14 regulates the expression of proteins critical for brain function. Utilizing this mouse model, future work will focus on characterizing specific target mRNAs, such as CaMK2a, which ZC3H14 could regulate to ensure proper brain function.
Materials and Methods
Generation of Zc3h14Δex13/Δex13 mouse
Mouse embryonic stem cells (clone EPD0366_5_F01, ES cell strain C57BL/6N, Parental Cell Line JM8A3.N1) were obtained from Knockout Mouse Project Repository (UC Davis, Davis, CA). This clone flanks with loxP sites exon 13, the first common exon among all four Zc3h14 isoforms (a, b, c, d) (see Fig. 1A and B). In vitro Flp-FRT recombination to eliminate the LacZ/neomycin cassette and subsequent blastocyst injections were performed by the Emory Mouse Transgenic and Gene Targeting Core. We generated heterozygous floxed Zc3h14 exon 13 mice (Zc3h14F/+) by mating adult male chimeras to C57BL/6N wildtype female mice (The Jackson Laboratory). Offspring generated from this cross were then mated to generate homozygous floxed mutant mice (Zc3h14F/F). EIIa-Cre mice (purchased from The Jackson Laboratory, Stock #003724, mixed C57BL/6J and C57BL/6N genetic background) which express Cre-recombinase active at the zygotic stage (29), were mated to homozygous floxed Zc3h14 exon 13 (Zc3h14F/F) mice for 3-4 generations to generate a germline-transmissible Zc3h14 allele lacking exon 13 (Zc3h14Δex13/Δex13). These mice with confirmed proper recombination were mated to wildtype C57BL/6 mice (purchased from The Jackson Laboratory) to breed out the EIIa-Cre allele. EIIa-Cre-negative, Zc3h14Δex13/+ were mated to generate Zc3h14Δex13/Δex13 mutant homozygous mice for at least four generations.
Control Zc3h14+/+ mice were maintained in the colony as control counterparts from the heterozygous Zc3h14Δex13/+ breeders. Generations F4-F8 of Zc3h14+/+ (and Zc3h14Δex13/Δex13) homozygous pairings were used for experiments, and homozygous off-spring were routinely cross-bred to minimize genetic drift between Zc3h14+/+ and Zc3h14Δex13/Δex13mice.
All procedures involving mice were done in accordance with the NIH guidelines for use and care of live animals and were approved by the Emory University Institutional Animal Care and Use Committee.
Genotyping
Mice were screened for the conditional floxed allele, Cre-mediated recombination, and the Cre-recombinase gene by PCR of genomic DNA isolated from toe clippings using a standard DNA isolation protocol (86). Tissue from toe clippings was lysed in 50 µl of standard lysis buffer supplemented with Proteinase K overnight at 55 °C. Samples were centrifuged for 5 min at full speed to sediment cellular debris. Genomic DNA was precipitated using ice-cold 100% ethanol, washed two times in 75% ethanol, and allowed to air dry before being resuspended in 30 µl of water. Template DNA was amplified by standard PCR per the Qiagen Taq DNA Polymerase PCR kit manufacturing protocol (Qiagen 201205). The following primers were used to genotype mice at the Zc3h14 locus (as illustrated in Fig. 1B and C): Fwd (5’-GTTGGCTCATCTTCTGTAAAGC-3’) and RevI (5’- GGTAAGGAAAACTAATCCACATCTAG-3’) for the conditional floxed allele (generating a product of either 885 bp for control, 994 bp for floxed, or 230 bp for recombined alleles) or RevII (5’-GCCACACTCAGGTCAGTCATCTCG-3’) for the direct detection of exon 13 (generating a product of either 469 bp for control or 625 bp for floxed, or no product for the recombined allele which excises exon 13). For detection of the Cre-recombinase gene, generic Cre-recombinase primers were used: Fwd-Cre (5’-GCGGTCTGGCAGTAAAAACTATC-3’) and Rev-Cre (5’GTGAAACAGCATTGCTGTCACTT-3’).
Immunoblotting
Mouse brain tissue samples were lysed in RIPA-2 buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% IGEPAL or NP-40, 0.5% deoxycholic acid, 0.1% SDS) containing protease inhibitors (Complete Mini; Roche) and centrifuged at 9,200 x g for 10 min at 4 °C. The pellet was discarded, and the supernatant was subjected to SDS-PAGE and immunoblotting. An equal amount of protein from each sample was loaded onto 10% SDS-polyacrylamide gels and transferred to a 0.2 μm nitrocellulose membrane (Bio-Rad Laboratories) (87). After blocking non-specific binding, the membranes were incubated with the primary antibodies. Primary antibodies used were as follows: anti-ZC3H14 (rabbit polyclonal antibody generated against the N-terminal PWI-like domain) (14), EIF5 (Santa Cruz sc-282), α-Tubulin (Sigma T9026), Histone H3 (Abcam ab1791), THOC1 (kind gift from Dr. Robin Reed, Harvard University, Boston, MA), S6 Ribosomal Protein (5G10) (Cell Signaling 2217), followed by incubation with the appropriate horseradish peroxidase (HRP)-conjugated secondary IgG antibody (Jackson ImmunoResearch).
Real-time PCR analysis of Zc3h14 splice variant d mRNA
To obtain RNA from mouse tissue, brain was lysed using QIAzol, according to the guidelines of the manufacturer (Qiagen). cDNA was synthesized from RNA isolated from mouse whole brain using M-MLV Reverse Transcriptase kit (Invitrogen 28025), GoTaq DNA Polymerase (Promega M3001), and Recombinant RNasin Ribonuclease Inhibitor (Promega N2511). Briefly, 5 ng of cDNA was subjected to PCR amplification using SYBR Green PCR Master Mix (Applied Biosystems) and Zc3h14 splice variant d-specific primers: Fwd (5’GGTTGAGAAAGGAACTCAACAGAGGC-3’) (junction of alternative exon 1 and exon 10) and Rev (5’-TCATCTCGGCTTGACTCATCTCCA-3’) (junction of exons 10 and 13). PCR amplification was performed with an Applied Biosystems, StepOnePlus Real-Time PCR System. The 18S rRNA was used to normalize mRNA. Relative quantitation of mRNA expression level was determined using the relative standard curve method as previously described (88) and according to the instructions of the manufacturer (Applied Biosystems).
ZC3H14 immunoprecipitation for mass spectrometry analysis
Brain tissue was homogenized in IP buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 32 mM NaF, 0.5% NP-40 in DEPC-treated water) containing protease inhibitors (Complete Mini; Roche; 1 tablet/10 ml of IP buffer). Cells were sonicated on ice 5 times at 0.5% output for 10 s, passed through a 27-gauge syringe 5 times, and placed back on ice for 20 min with occasional vortexing. Lysates were spun at 15,600 × g for 10 min at 4 °C and protein concentrations were determined with a standard BCA assay. Protein G magnetic beads (DYNA beads, Invitrogen) were rinsed and resuspended in IP buffer and incubated with pre-immune rabbit serum or an equal volume of N-terminal ZC3H14 antibody for 1 h at room temperature. Bead/antibody and bead/pre-immune samples were rinsed in IP buffer and added to clarified cell lysates. Ten percent of the sample was then removed and reserved for input analysis, and the remainder of the sample was incubated overnight at 4 °C while tumbling end over end. Beads were then magnetized to collect bound and unbound species separately. The beads were washed 5 times with ice-cold IP buffer. ZC3H14 protein complexes were eluted with reducing sample buffer (RSB): 250 mM Tris HCL, 500 mM DTT, 10% SDS, 0.5% bromophenol blue, 50% glycerol. Upon submission to the Emory Proteomics Core for mass spectrometry analysis, the samples were further reduced with DTT and alkylated with Iodoacetamide (IAA), and then subjected to sequential in-solution digestion with LysC (1:100 enzyme-to-protein ratio) for 4 hrs and with trypsin (1:100 enzyme-to-protein ratio) overnight. After desalting and sample cleanup with Water’s Sep-Pak, the samples were submitted for analysis on the Orbitrap Fusion Mass Spectrometer (Thermo Scientific).
Immunohistochemical analysis of brain morphology
Whole brains were fixed overnight in 4% PFA, processed for routine embedding in paraffin, and cut 9 μm in thickness onto glass slides. Slides were dewaxed in xylene and rehydrated through graded ethanol washes. Haematoxylin and eosin staining was performed using standard procedures. Alternatively, slices were stained with cresyl violet to assess hippocampal morphology (89).
Images were visualized on an Olympus BX51 microscope using a 2X magnification objective and captured using DP Controller (Olympus) software. Genotype of all brain images was blinded for the analyses, and the quantification of ventricular area was performed using ImageJ.
Behavioral and function and coordination assays
For all behavioral assays, the investigator was blinded to the genotype of the mice. All studies were performed with independent cohorts of mice.
Y maze
The Y-maze is made of three centrally-joined arms (8 in-tall × 15.5 in-long × 4 in-wide) made of opaque Plexiglas with an arm angle of 120°. Using 10 Zc3h14+/+and 10 Zc3h14Δex13/Δex13male mice, mice were individually placed in the central intersection and allowed to explore the maze for 8 min. Arm entries were automatically recorded using TopScan software by CleverSys. The mouse’s midpoint needed to be inside the arm to count as an entry. Correct alternation is defined as entry into an arm different from the last two arm entries (e.g. ABC, CAB, BCA, CBA, BAC, or ACB, but not ABA, CAC, etc.). The maze was cleaned with Virkon between each test to remove olfactory cues. % correct alternation was calculated as: % Correct Alternation= [(Number of Correct Alternations)/(Total arm entries)] x 100. The Y-maze assay was performed by Jason Schroeder with the Emory University Rodent Behavioral Core.
Fear conditioning
Fear conditioning was performed using an established Coulbourn system (7 × 7 × 12 in), utilizing a metal shock grid floor. On the training day, mice were allowed to acclimate to the chamber for 3 min followed by three trials of tone (85 dB)-shock (0.5 mA) pairings. Each tone-shock session lasted 20 s, with the shock occurring within the last 0.5 s of the tone. Each trial was separated by 1 min. To assay contextual freezing, mice were returned to the same chamber the following day for 8 min with no tone or shocks administered and measured for percent freezing. To assay cued fear, mice were placed in a new chamber the next day for 3 min, followed by the 85 dB tone for 6 min and measured for percent freezing. Percent freezing (defined as a lack of any movement/total time x 100) was measured each day in 1-min intervals (FreezeFrame software; Actimetrics). The Fear Conditioning assay was performed by Jason Schroeder with the Emory University Rodent Behavioral Core.
Water radial arm maze (WRAM)
The WRAM apparatus was used as previously described (41). A radial maze, 2 m in diameter with 8 arms of equal width and length (10 cm wide and 60 cm long) at 45-degree angles surrounding an open center section, was filled with water at approximately 21 °C (Fig. 4C). The water was made opaque by mixing in water-soluble, non-toxic paint of the same color (black) as the maze and the platforms. The water was filled to a level 0.5–1.0 cm above the top of all four total submerged platforms, placed one each at the far end of half of the arms. A specific orientation of platforms, constant for each mouse throughout the testing period, was placed in the arena with the following criteria: the ‘start’ arm always contained no platform and no more than 2 adjacent arms contained platforms.
Ten Zc3h14+/+ and nine Zc3h14Δex13/Δex13 male adult (5-month-old) mice were tested. At the start of each trial, a mouse was placed inside a clean, dry, empty cage heated with a lamp to approximately 40 °C. Once the platforms were placed in the correct orientation, the mouse was gently placed in the water at the far end of the ‘start’ arm, and the timer was started. A blinded researcher monitored the progress of the mouse by recording both the sequence of arms the mouse entered and the time in seconds the mouse took to locate a platform, with a maximum trial length of 120 s before the trial was aborted and the mouse was gently guided to the nearest platform. When a mouse reached a platform, it was allowed to remain there for 15 s, allowing time for it to identify any visual cues placed on the walls to aid in spatial memory. The mouse was then returned to the heated and dry cage for a period of 30 s as a reward for successfully completing the trial. The platform on which the mouse was seated was removed, and the next trial commenced in the same manner. Four trials were performed per mouse, per day, until each mouse had located all 4 platforms. Testing was conducted for 9 consecutive days.
We used the following criteria to analyse WRAM data. To plot total latency, time required to reach the platforms were recorded for each mouse and summed across the four trials per day. After the groups were unblinded, these latencies were calculated across the 9-day testing period. To plot overall total errors, each entry into an arm that did not result in locating a platform was summed across the four trials for each mouse per day. These errors were then sorted into 3 types: I) errors in which the first time on a given trial a mouse enters any arm that contained a platform on a previous trial on the same day (shows good working and spatial memory but poor procedural memory); II) errors in which the first time on a given trial a mouse enters any arm that never contained a platform or else enters an arm with a platform but does not successfully locate the platform (shows poor spatial memory); III) errors in which a mouse makes a repeated entry into any arm for which it has previously registered a type I or type II error on the same trial (shows poor working memory). To plot working memory errors, groups were then unblinded, and type III errors were summed for the four trials per day across the 9-day testing period, at which point a steady-state minimum was achieved, indicating sufficient performance. Sufficient performance on the WRAM requires that a group makes an average of no more than approximately 1.0 type III error per subject per day by the conclusion of testing in order to demonstrate learning.
Open field
Open field was performed as previously described (90). Briefly, the mice were acclimated to the testing room for 2 h prior to initiating the task. Twelve Zc3h14+/+ and twelve Zc3h14Δex13/Δex13male adult mice (3–4 months) were examined. Each mouse was placed into the center of an opaque Plexiglas box (60 × 60 × 60 cm) and given 10 min to explore the apparatus. The center zone was defined as the region 15 cm from each side of the box. The time spent in the center, number of entries into the center, total distance traveled, and average speed were recorded and scored with AnyMaze video tracking software (Stoelting Co. IL) by a user blinded to genotype.
Novel cage
Twelve Zc3h14+/+ and twelve Zc3h14Δex13/Δex13male adult mice (3–4 months) were examined. Each mouse was placed into an uncovered cage with corncob bedding for 10 min. Exploratory actions such as rearing, digging, and grooming were scored.
Light-dark box
Twelve Zc3h14+/+ and twelve Zc3h14Δex13/Δex13male adult mice (3–4 months) were examined. The apparatus comprised a Plexiglas box (30 × 14 × 14.5 cm) that was divided into a light (20 × 14 × 14.5 cm) and dark side (10 × 14 × 14.5 cm). The walls of the dark side were blackened to reduce external illumination. A hole in the divider between the two sides enabled movement of the mouse between the two sections. Each mouse was placed into the light side and given 10 min to explore freely. Latencies to first enter the dark side, the number of transitions between each side, and total time spent in each side was recorded for each mouse.
Wire-hang assay
Testing was performed with 1- × 1-cm mesh hardware fixed to a 19- × 19-cm frame. Nine Zc3h14+/+ and 14 Zc3h14Δex13/Δex13 male adult mice were placed on the wire mesh, which was slowly inverted 27.5 cm over a soft, thick heating pad (unheated), lined with paper towels. Latency to fall was recorded in seconds as previously described (91). The test was performed three trials a day for three days for each mouse. The maximum latency to fall for each day was averaged together for the three days for each mouse.
Rotarod
A four-lane motorized rotarod (Rotamex-5 Controller, Columbus Instruments, Columbus, OH) was used according to manufacturer’s instructions and as previously described (91). Each rod was 3.0 cm in diameter and 9.5 cm long. The fall height from rod center was 44.5 cm to the base. Scanning infrared beam sensors monitored animals’ absence from rod to detect time to fall. The acceleration interval (1.0 s), acceleration step (0.1 RPM), and maximum speed (50.0 RPM) was the same for each mouse and for all testing days. Prior to testing, mice were trained on a rotarod (AccuRotor Rota Rod RR4/M, AccuScan Instruments, Columbus, OH) that had a constant speed of 4.2 RPM.
Nine Zc3h14+/+ and 10 Zc3h14Δex13/Δex13 male adult mice were allowed to acclimate to the room for 15 min. All mice were oriented on the rod to walk forward to maintain balance. To avoid falling, mice were required to move forward in a coordinated manner. Training occurred over 3 days. Four mice at a time were trained for 10 min on the rotarod at constant speed, replacing mice back on the rod when subjects had fallen from the rotarod. After training on the third day, the mice started the 3-day testing phase. Each day of testing consisted of 3 trials, with at least 15 min of rest in between trials, for a total of 9 trials. Latency to fall was averaged for the three trials within a particular day and analysed using the two-tailed Student's t-test with the Bonferroni correction for repeated measures. P values of 0.05 or less were taken as statistically significant. The experimenter was blinded to the genotype of the mice until all latency times were recorded.
Optokinetic tracking (OKT)
To assess visual function, we measured visual acuity and contrast sensitivity using optokinetic tracking behavior (OptoMotry, Cerebral Mechanics, Inc., Lethbridge, Alberta, Canada) (92–94). Both mice employed for the WRAM analyses and independent cohorts were assessed in these assays. Four Zc3h14+/+ and four Zc3h14Δex13/Δex13 mice were placed on a platform in the center of a virtual optomotor drum consisting of four computer monitors. Spatial frequency thresholds were determined by varying the spatial frequency at 100% contrast. Contrast sensitivity thresholds were determined by varying contrast at peak contrast sensitivity (0.064 cycles/degree). Positive responses were noted as reflexive head-tracking in the direction of the moving gratings. Both eyes were tested by using clockwise and counter-clockwise rotations (43). The values from the left and right eyes were averaged for analysis of differences between groups at 6 weeks of age using a two-way repeated ANOVA with Holm-Sidak post-hoc.
Determination of bulk poly(A) tail length
Bulk poly(A) tails were analysed using a standard poly(A) tail length assay (95) as described previously (96). Total RNA was isolated from hippocampi of P0 Zc3h14+/+ and Zc3h14Δex13/Δex13mice using a RNeasy mini kit (Qiagen 74104). Total RNA was 3’end labeled with [32P]pCp using T4 RNA ligase and digested with a cocktail of RNAse A/T1. The resulting tracts of poly(A) RNA were resolved by gel electrophoresis and imaged by film or PhosphorImager. Three biological replicates were performed for each mouse genotype analysed. Using ImageJ software, densitometry analysis of each experiment was performed in order to obtain profile curves for each sample that was analysed, as described previously (12). Profiles of the relative signal intensity at a given location on the gel were plotted in Microsoft Excel and a moving average (period = 25) calculated. For all analysis, statistical significance was determined using a two-tailed Mann Whitney test.
Mass spectrometry analysis of Zc3h14+/+ and Zc3h14Δex13/Δex13 hippocampi
Samples submitted for mass spectrometry analysis consisted of eight hippocampi from four Zc3h14+/+ and eight hippocampi from four Zc3h14Δex13/Δex13 mice at P0. Each tissue piece was uniformly homogenized in 300 µl of urea lysis buffer (8 M urea, 100 mM NaHPO4, pH 8.5), including 3 µl (100x stock) HALT protease and phosphatase inhibitor cocktail (Pierce). All homogenization was performed using a Bullet Blender (Next Advance) according to manufacturer protocols. Briefly, each tissue piece was added to Urea lysis buffer in a 1.5 ml Rino tube (Next Advance) harboring 750 mg stainless steel beads (0.9–2 mm in diameter) and blended twice for 5-min intervals in the cold room (4 °C). Protein supernatants were transferred to 1.5 ml Eppendorf tubes and sonicated (Sonic Dismembrator, Fisher Scientific) 3 times for 5 s with 15 s intervals of rest at 30% amplitude to disrupt nucleic acids and subsequently vortexed. Protein concentration was determined by the bicinchoninic acid (BCA) method, and samples were frozen in aliquots at −80 °C. Protein homogenates (100 µg) were diluted with 50 mM NH4HCO3 to a final concentration of less than 2 M urea and then treated with 1 mM dithiothreitol (DTT) at 25 °C for 30 min, followed by 5 mM iodoacetimide (IAA) at 25 °C for 30 min in the dark. Protein was digested with 1:100 (w/w) lysyl endopeptidase (Wako) at 25 °C for 2 h and further digested overnight with 1:50 (w/w) trypsin (Promega) at 25 °C. Resulting peptides were desalted with a Sep-Pak C18 column (Waters) and dried under vacuum.
For LC-MS/MS analysis, hippocampal derived peptides were resuspended in peptide 100 µl of loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile). Peptide mixtures (2 µl) were separated on a self-packed C18 (1.9 µm Dr. Maisch, Germany) fused silica column (25 cm × 75 µM internal diameter (ID); New Objective, Woburn, MA) by a Dionex Ultimate 3000 RSLCNano and monitored on a Fusion mass spectrometer (ThermoFisher Scientific, San Jose, CA). Elution was performed over a 120-min gradient at a rate of 400 nl/min with buffer B ranging from 3 to 80% (buffer A: 0.1% formic acid in water, buffer B: 0.1% formic in acetonitrile). The mass spectrometer cycle was programmed to collect at the top speed for 3 s cycles. The MS scans (400–1600 m/z range, 200,000 AGC, 50 ms maximum ion time) were collected at a resolution of 120,000 at m/z 200 in profile mode and the HCD MS/MS spectra (0.7 m/z isolation width, 30% collision energy, 10,000 AGC target, 35 ms maximum ion time) were detected in the ion trap. Dynamic exclusion was set to exclude previous sequenced precursor ions for 20 s within a 10 ppm window. Precursor ions with +1, and +8 or higher charge states were excluded from sequencing.
MaxQuant for label-free quantification
Raw data files were analysed using MaxQuant v1.5.2.8 with Thermo Foundation 2.0 for RAW file reading capability. The search engine Andromeda was used to build and search a concatenated target-decoy IPI/Uniprot mouse reference (downloaded at Aug 14, 2015) supplement with eGFP protein sequence. Protein Methionine oxidation (+15.9949 Da) and protein N-terminal acetylation (+42.0106 Da) were variable modifications (up to 5 allowed per peptide); cysteine was assigned a fixed carbamidomethyl modification (+57.0215 Da). Only fully tryptic peptides were considered with up to 2 miscleavages in the database search. A precursor mass tolerance of ±10 ppm was applied prior to mass accuracy calibration and ±4.5 ppm after internal MaxQuant calibration. Other search settings included a maximum peptide mass of 6,000 Da, a minimum peptide length of 6 residues, 0.6 Da Tolerance for iron trap HCD MS/MS scans. The false discovery rate (FDR) for peptide spectral matches, proteins, and site decoy fraction were all set to 1%. The label free quantitation (LFQ) algorithm in MaxQuant (44,97) was used for protein quantitation. LFQ intensity of each protein for each mouse was averaged from two hippocampi (left and right). No more than two missing values were considered per group (Zc3h14Δex13/Δex13 or Zc3h14+/+). Differentially expressed proteins were found by calculating Student’s t-test P values and fold difference |log2(Zc3h14Δex13/Δex13/Zc3h14+/+)| ≥ 0.32 (≥ ±1.25-fold change). Volcano plots were plotted with ggplot2 packages in R.
GO enrichment, network and heatmap
Functional enrichment of the modules was determined using the GO-Elite (v1.2.5) package (98). The set of total proteins identified and quantified (n = 4,161) was used as the background. Input lists included proteins either significantly decreased (n = 50) or increased (n = 63) in Zc3h14Δex13/Δex13 mice. Z-score determines overrepresentation of ontologies in a module and permutation P-value was used to assess the significance of the Z-score. For these down-regulated proteins in Zc3h14Δex13/Δex13 mice, Z-score cut off of 1.96, P value cut off of 0.05 with a minimum of 3 proteins per category were used as filters in pruning the ontologies. For these up regulated proteins in Zc3h14Δex13/Δex13mice, Z-score cut off of 2.58, P value cut off of 0.01 with a minimum of 5 proteins per category were used as filters in pruning the ontologies. Horizontal bar graph was plotted in R. The networks were constructed using the igraph package in R (99). The heatmap and hierarchical clustering were generated using the default parameters in Perseus (100).
Statistical analysis
Data were analysed using R, GraphPad Prism 6.0, or Microsoft Excel. R, Prism, or Microsoft Excel was also used to generate graphs, column and row means and standard error of the means (SEMs), and statistical analyses (two-way ANOVA, t-tests, and post-hoc analysis) to compare between genotypes. P values are > 0.05, unless otherwise noted. P values < 0.05 were considered statistically significant. Error bars indicate SEM unless otherwise noted.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to W. Rossoll for technical assistance in locating the mouse genome sequence and providing early input to establish the Zc3h14 mouse model, K. R. Williams for technical assistance in primary neuronal culture, A. L. Mattheyses and L. Fox-Gooharioon for instruction on confocal microscope imaging, A. Rojas for thoughtful suggestions on histological analysis, the William Tyor lab for providing the WRAM apparatus, S. Yang and X. J. Li for assistance in using and providing the rotarod machine, D. J. Cutler for statistical guidance, and A. Beain for help with genotyping. Special thanks to the following individuals of Emory University core facilities: H. Zhang and T. Caspary of the Mouse Transgenic and Gene Targeting Core for blastocyst injection and production of chimeric mice, E. Dammer and D. Duong of the Proteomics Core for mass spectrometry analysis, M. Gearing and D. S. Cooper at the Center for Neurodegenerative Diseases Histopathology Core for processing and staining tissues, and Division of Animal Resources, particularly C. Pitt, K. Alford, and D. Amankwah, for daily mouse colony maintenance. We appreciate thoughtful guidance and mentorship on work with mouse models from K. Ressler, Y. Feng, M. Ifrim, V. H. Haase, and E. B. Rankin.
Conflict of Interest statement. None declared.
Funding
Emory University Integrated Cellular Imaging Microscopy Core of the Emory and Children's Pediatric Research Center. Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Gates Millennium Scholars (to J.R.), VA Career Development Award BX001677 (to P.S.G.), James S. McDonnell Foundation 22020346 (to J.F. and P.S.G.), and the following National Institutes of Health grants: F31HD070735 (to J.R.), P20NS055077 (to J. R. for imaging), 1R21MH105353 (to G.M.B. and C.G.), R01NS072221 (to A.E.), R01MH10730501 (to A.H.C. and K.H.M.), R01GM05872815 and R21AG054206 (to A.H.C).
References
- 1. Moore M.J. (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science, 309, 1514–1518. [DOI] [PubMed] [Google Scholar]
- 2. Shyu A.B., Wilkinson M.F., van Hoof A. (2008) Messenger RNA regulation: to translate or to degrade. embo J., 27, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neelamraju Y., Hashemikhabir S., Janga S.C. (2015) The human RBPome: From genes and proteins to human disease. J. Proteomics, 127, 61–70. [DOI] [PubMed] [Google Scholar]
- 4. Lukong K.E., Chang K.W., Khandjian E.W., Richard S. (2008) RNA-binding proteins in human genetic disease. Trends Genet., 24, 416–425. [DOI] [PubMed] [Google Scholar]
- 5. Dredge B.K., Polydorides A.D., Darnell R.B. (2001) The splice of life: alternative splicing and neurological disease. Nat. Rev. Neurosci., 2, 43–50. [DOI] [PubMed] [Google Scholar]
- 6. Squire L.R., Kandel E.R. (2000) Memory: From Mind to Molecules. Scientific American Library, New York. [Google Scholar]
- 7. Lenzken S.C., Achsel T., Carri M.T., Barabino S.M. (2014) Neuronal RNA-binding proteins in health and disease. Wiley Interdiscip. Rev. RNA, 5, 565–576. [DOI] [PubMed] [Google Scholar]
- 8. Xing L., Bassell G.J. (2013) mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic, 14, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santoro M.R., Bray S.M., Warren S.T. (2012) Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol., 7, 219–245. [DOI] [PubMed] [Google Scholar]
- 10. Pak C., Garshasbi M., Kahrizi K., Gross C., Apponi L.H., Noto J.J., Kelly S.M., Leung S.W., Tzschach A., Behjati F.. et al. (2011) Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc. Natl Acad. Sci. U S A, 108, 12390–12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly S.M., Bienkowski R., Banerjee A., Melicharek D.J., Brewer Z.A., Marenda D.R., Corbett A.H., Moberg K.H. (2016) The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Develop. Neurobiol., 76, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelly S.M., Leung S.W., Pak C., Banerjee A., Moberg K.H., Corbett A.H. (2014) A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. rna, 20, 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelly S.M., Pabit S.A., Kitchen C.M., Guo P., Marfatia K.A., Murphy T.J., Corbett A.H., Berland K.M. (2007) Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl Acad. Sci., 104, 12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leung S.W., Apponi L.H., Cornejo O.E., Kitchen C.M., Valentini S.R., Pavlath G.K., Dunham C.M., Corbett A.H. (2009) Splice variants of the human ZC3H14 gene generate multiple isoforms of a zinc finger polyadenosine RNA binding protein. Gene, 439, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anderson J.T., Wilson S.M., Datar K.V., Swanson M.S. (1993) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell Biol., 13, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fasken M.B., Stewart M., Corbett A.H. (2008) Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J. Biol. Chem., 283, 27130–27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelly S.M., Leung S.W., Apponi L.H., Bramley A.M., Tran E.J., Chekanova J.A., Wente S.R., Corbett A.H. (2010) Recognition of Polyadenosine RNA by the Zinc Finger Domain of Nuclear Poly(A) RNA-binding Protein 2 (Nab2) Is Required for Correct mRNA 3′-End Formation*. J. Biol. Chem., 285, 26022–26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelly S., Pak C., Garshasbi M., Kuss A., Corbett A.H., Moberg K. (2012) New kid on the ID block: neural functions of the Nab2/ZC3H14 class of Cys3His tandem zinc-finger polyadenosine RNA binding proteins. RNA Biol., 9, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhlmann S.I., Valkov E., Stewart M. (2014) Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers. Nucleic Acids Res., 42, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guthrie C.R., Greenup L., Leverenz J.B., Kraemer B.C. (2011) MSUT2 is a determinant of susceptibility to tau neurotoxicity. Hum. Mol. Genet., 20, 1989–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reuter L.M., Meinel D.M., Sträßer K. (2015) The poly(A)-binding protein Nab2 functions in RNA polymerase III transcription. Genes Dev., 29, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hector R.E., Nykamp K.R., Dheur S., Anderson J.T., Non P.J., Urbinati C.R., Wilson S.M., Minvielle-Sebastia L., Swanson M.S. (2002) Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. embo J., 21, 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soucek S., Zeng Y., Bellur D.L., Bergkessel M., Morris K.J., Deng Q., Duong D., Seyfried N.T., Guthrie C., Staley J.P.. et al. (2016) The Evolutionarily-conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre-mRNA. Mol Cell Biol, pii: MCB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marfatia K.A., Crafton E.B., Green D.M., Corbett A.H. (2003) Domain Analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. J. Biol. Chem., 278, 6731–6740. [DOI] [PubMed] [Google Scholar]
- 25. Lee D.C., Aitchison J.D. (1999) Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem., 274, 29031–29037. [DOI] [PubMed] [Google Scholar]
- 26. Duncan K., Umen J.G., Guthrie C. (2000) A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol., 10, 687–696. [DOI] [PubMed] [Google Scholar]
- 27. Green D.M., Marfatia K.A., Crafton E.B., Zhang X., Cheng X., Corbett A.H. (2002) Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem., 277, 7752–7760. [DOI] [PubMed] [Google Scholar]
- 28. Brockmann C., Soucek S., Kuhlmann S.I., Mills-Lujan K., Kelly S.M., Yang J.C., Iglesias N., Stutz F., Corbett A.H., Neuhaus D.. et al. (2012) Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure, 20, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl Acad. Sci. U S A, 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dooley T.P., Miranda M., Jones N.C., DePamphilis M.L. (1989) Transactivation of the adenovirus EIIa promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development, 107, 945–956. [DOI] [PubMed] [Google Scholar]
- 31. Abdeen S.K., Del Mare S., Hussain S., Abu-Remaileh M., Salah Z., Hagan J., Rawahneh M., Pu X.A., Russell S., Stein J.L.. et al. (2013) Conditional inactivation of the mouse Wwox tumor suppressor gene recapitulates the null phenotype. J. Cell Physiol., 228, 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerber E.E., Gallo E.M., Fontana S.C., Davis E.C., Wigley F.M., Huso D.L., Dietz H.C. (2013) Integrin-modulating therapy prevents fibrosis and autoimmunity in mouse models of scleroderma. Nature, 503, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y., Ju T., Ding X., Xia B., Wang W., Xia L., He M., Cummings R.D. (2010) Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl Acad. Sci. U S A, 107, 9228–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao Y., Pahlberg J., Sarria I., Kamasawa N., Sampath A.P., Martemyanov K.A. (2012) Regulators of G protein signaling RGS7 and RGS11 determine the onset of the light response in ON bipolar neurons. Proc. Natl Acad. Sci. U S A, 109, 7905–7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chandra D., Jia F., Liang J., Peng Z., Suryanarayanan A., Werner D.F., Spigelman I., Houser C.R., Olsen R.W., Harrison N.L.. et al. (2006) GABAA receptor alpha 4 subunits mediate extrasynaptic inhibition in thalamus and dentate gyrus and the action of gaboxadol. Proc. Natl Acad. Sci. U S A, 103, 15230–15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergo M.O., Gavino B., Ross J., Schmidt W.K., Hong C., Kendall L.V., Mohr A., Meta M., Genant H., Jiang Y.. et al. (2002) Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl Acad. Sci. U S A, 99, 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Higashi Y., Maruhashi M., Nelles L., Van de Putte T., Verschueren K., Miyoshi T., Yoshimoto A., Kondoh H., Huylebroeck D. (2002) Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis, 32, 82–84. [DOI] [PubMed] [Google Scholar]
- 38. Deacon R.M., Rawlins J.N. (2006) T-maze alternation in the rodent. Nat. Protoc., 1, 7–12. [DOI] [PubMed] [Google Scholar]
- 39. Vorhees C.V., Williams M.T. (2014) Assessing spatial learning and memory in rodents. ILAR J., 55, 310–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bunge S.A., Ochsner K.N., Desmond J.E., Glover G.H., Gabrieli J.D. (2001) Prefrontal regions involved in keeping information in and out of mind. Brain, 124, 2074–2086. [DOI] [PubMed] [Google Scholar]
- 41. Penley S.C., Gaudet C.M., Threlkeld S.W. (2013) Use of an eight-arm radial water maze to assess working and reference memory following neonatal brain injury. J. Vis. Exp., 4, 50940.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morris R.G., Garrud P., Rawlins J.N., O'Keefe J. (1982) Place navigation impaired in rats with hippocampal lesions. Nature, 297, 681–683. [DOI] [PubMed] [Google Scholar]
- 43. Douglas R.M., Alam N.M., Silver B.D., McGill T.J., Tschetter W.W., Prusky G.T. (2005) Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis. Neurosci., 22, 677–684. [DOI] [PubMed] [Google Scholar]
- 44. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shonesy B.C., Jalan-Sakrikar N., Cavener V.S., Colbran R.J. (2014) CaMKII: a molecular substrate for synaptic plasticity and memory. Prog. Mol. Biol. Transl Sci., 122, 61–87. [DOI] [PubMed] [Google Scholar]
- 46. Chang H.T., Liu C.C., Chen S.T., Yap Y.V., Chang N.S., Sze C.I. (2014) WW domain-containing oxidoreductase in neuronal injury and neurological diseases. Oncotarget, 5, 11792–11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bienkowski R., Banerjee A., Rounds J.C., Rha J., Omotade O.F., Gross C., Morris K.J., Pak C., Jones S.K., Santoro M.R.. et al. (2017) The conserved, disease-associated RNA binding protein dNab2 interacts with the Fragile-X protein ortholog in Drosophila neurons. Cell Rep.., submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wigington C.P., Morris K.J., Newman L.E., Corbett A.H. (2016) The polyadenosine RNA-binding protein, zinc finger Cys3His protein 14 (ZC3H14), regulates the pre-mRNA processing of a key ATP synthase subunit mRNA. J. Biol. Chem., 291, 22442–22459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou H., Mangelsdorf M., Liu J., Zhu L., Wu J.Y. (2014) RNA-binding proteins in neurological diseases. Sci. China Life Sci., 57, 432–444. [DOI] [PubMed] [Google Scholar]
- 50. Kapeli K., Yeo G.W. (2012) Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. Front. Neurosci., 6, 144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paronetto M.P., Sette C. (2010) Role of RNA-binding proteins in mammalian spermatogenesis. Int. J. Androl., 33, 2–12. [DOI] [PubMed] [Google Scholar]
- 52. Venables J.P., Eperon I. (1999) The roles of RNA-binding proteins in spermatogenesis and male infertility. Curr. Opin. Genet. Dev., 9, 346–354. [DOI] [PubMed] [Google Scholar]
- 53. Cheng C.Y., Wu J.C., Tsai J.W., Nian F.S., Wu P.C., Kao L.S., Fann M.J., Tsai S.J., Liou Y.J., Tai C.Y.. et al. (2015) ENU mutagenesis identifies mice modeling Warburg Micro Syndrome with sensory axon degeneration caused by a deletion in Rab18. Exp. Neurol., 267, 143–151. [DOI] [PubMed] [Google Scholar]
- 54. Bakker N.E., Wolffenbuttel K.P., Looijenga L.H., Hokken-Koelega A.C. (2015) Testes in infants with Prader-Willi syndrome: human chorionic gonadotropin treatment, surgery and histology. J. Urol., 193, 291–298. [DOI] [PubMed] [Google Scholar]
- 55. Sammour Z.M., Gomes C.M., de Bessa J. Jr., Pinheiro M.S., Kim C.A., Hisano M., Bruschini H., Srougi M. (2014) Congenital genitourinary abnormalities in children with Williams-Beuren syndrome. J. Pediatr. Urol., 10, 804–809. [DOI] [PubMed] [Google Scholar]
- 56. Yi T., Weng J., Siwko S., Luo J., Li D., Liu M. (2014) LGR4/GPR48 inactivation leads to aniridia-genitourinary anomalies-mental retardation syndrome defects. J. Biol. Chem., 289, 8767–8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cabezas D.A., Slaugh R., Abidi F., Arena J.F., Stevenson R.E., Schwartz C.E., Lubs H.A. (2000) A new X linked mental retardation (XLMR) syndrome with short stature, small testes, muscle wasting, and tremor localises to Xq24-q25. J. Med. Genet., 37, 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siffroi J.P., Pawlak A., Alfonsi M.F., Troalen F., Guellaen G., Dadoune J.P. (2001) Expression of the TAR RNA binding protein in human testis. Mol. Hum. Reprod., 7, 219–225. [DOI] [PubMed] [Google Scholar]
- 59. Kimura M., Ishida K., Kashiwabara S., Baba T. (2009) Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol. Reprod., 80, 545–554. [DOI] [PubMed] [Google Scholar]
- 60. Sutherland J.M., Siddall N.A., Hime G.R., McLaughlin E.A. (2015) RNA binding proteins in spermatogenesis: an in depth focus on the Musashi family. Asian J. Androl., 17, 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Heisenberg M. (2003) Mushroom body memoir: from maps to models. Nat. Rev. Neurosci., 4, 266–275. [DOI] [PubMed] [Google Scholar]
- 62. Michel C.I., Kraft R., Restifo L.L. (2004) Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J. Neurosci., 24, 5798–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reiss A.L., Abrams M.T., Greenlaw R., Freund L., Denckla M.B. (1995) Neurodevelopmental effects of the FMR-1 full mutation in humans. Nat. Med., 1, 159–167. [DOI] [PubMed] [Google Scholar]
- 64. Ishihara K., Amano K., Takaki E., Shimohata A., Sago H., Epstein C.J., Yamakawa K. (2010) Enlarged brain ventricles and impaired neurogenesis in the Ts1Cje and Ts2Cje mouse models of Down syndrome. Cereb. Cortex, 20, 1131–1143. [DOI] [PubMed] [Google Scholar]
- 65. Correa-Cerro L.S., Wassif C.A., Kratz L., Miller G.F., Munasinghe J.P., Grinberg A., Fliesler S.J., Porter F.D. (2006) Development and characterization of a hypomorphic Smith-Lemli-Opitz syndrome mouse model and efficacy of simvastatin therapy. Hum. Mol. Genet., 15, 839–851. [DOI] [PubMed] [Google Scholar]
- 66. White N.S., Alkire M.T., Haier R.J. (2003) A voxel-based morphometric study of nondemented adults with Down Syndrome. Neuroimage, 20, 393–403. [DOI] [PubMed] [Google Scholar]
- 67. Pearlson G.D., Breiter S.N., Aylward E.H., Warren A.C., Grygorcewicz M., Frangou S., Barta P.E., Pulsifer M.B. (1998) MRI brain changes in subjects with Down syndrome with and without dementia. Dev. Med. Child Neurol., 40, 326–334. [PubMed] [Google Scholar]
- 68. Driscoll I., Davatzikos C., An Y., Wu X., Shen D., Kraut M., Resnick S.M. (2009) Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology, 72, 1906–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eckmann C.R., Rammelt C., Wahle E. (2011) Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA, 2, 348–361. [DOI] [PubMed] [Google Scholar]
- 70. Curinha A., Oliveira Braz S., Pereira-Castro I., Cruz A., Moreira A. (2014) Implications of polyadenylation in health and disease. Nucleus, 5, 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Darnell J.C., Richter J.D. (2012) Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb. Perspect. Biol., 4, a012344.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu-Yesucevitz L., Bassell G.J., Gitler A.D., Hart A.C., Klann E., Richter J.D., Warren S.T., Wolozin B. (2011) Local RNA translation at the synapse and in disease. J. Neurosci., 31, 16086–16093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Richter J.D., Bassell G.J., Klann E. (2015) Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci., 16, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Udagawa T., Farny N.G., Jakovcevski M., Kaphzan H., Alarcon J.M., Anilkumar S., Ivshina M., Hurt J.A., Nagaoka K., Nalavadi V.C.. et al. (2013) Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat. Med., 19, 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weill L., Belloc E., Bava F.A., Mendez R. (2012) Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol., 19, 577–585. [DOI] [PubMed] [Google Scholar]
- 76. Richter J.D. (2007) CPEB: a life in translation. Trends Biochem. Sci., 32, 279–285. [DOI] [PubMed] [Google Scholar]
- 77. Kreitz C., Furley P., Memmert D., Simons D.J. (2015) Working-memory performance is related to spatial breadth of attention. Psychol. Res., 79, 1034–1041. [DOI] [PubMed] [Google Scholar]
- 78. Rummel J., Boywitt C.D. (2014) Controlling the stream of thought: working memory capacity predicts adjustment of mind-wandering to situational demands. Psychon. Bull. Rev., 21, 1309–1315. [DOI] [PubMed] [Google Scholar]
- 79. Kail R.V., Lervag A., Hulme C. (2015) Longitudinal evidence linking processing speed to the development of reasoning. Dev Sci, 19, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nagel I.E., Chicherio C., Li S.C., von Oertzen T., Sander T., Villringer A., Heekeren H.R., Backman L., Lindenberger U. (2008) Human aging magnifies genetic effects on executive functioning and working memory. Front. Hum. Neurosci., 2, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kazdoba T.M., Leach P.T., Silverman J.L., Crawley J.N. (2014) Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res., 3, 118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liao L., Park S.K., Xu T., Vanderklish P., Yates J.R. 3rd (2008) Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc. Natl Acad. Sci. U S A, 105, 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Klemmer P., Meredith R.M., Holmgren C.D., Klychnikov O.I., Stahl-Zeng J., Loos M., van der Schors R.C., Wortel J., de Wit H., Spijker S.. et al. (2011) Proteomics, ultrastructure, and physiology of hippocampal synapses in a fragile X syndrome mouse model reveal presynaptic phenotype. J. Biol. Chem., 286, 25495–25504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang B., Wang T., Wan H., Han L., Qin X., Zhang Y., Wang J., Yu C., Berton F., Francesconi W.. et al. (2015) Fmr1 deficiency promotes age-dependent alterations in the cortical synaptic proteome. Proc. Natl Acad. Sci. U S A, 112, E4697–E4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sudhakaran I.P., Hillebrand J., Dervan A., Das S., Holohan E.E., Hulsmeier J., Sarov M., Parker R., VijayRaghavan K., Ramaswami M. (2014) FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl Acad. Sci. U S A, 111, E99–E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Laird P.W., Zijderveld A., Linders K., Rudnicki M.A., Jaenisch R., Berns A. (1991) Simplified mammalian DNA isolation procedure. Nucleic Acids Res., 19, 4293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Simionescu-Bankston A., Leoni G., Wang Y., Pham P.P., Ramalingam A., DuHadaway J.B., Faundez V., Nusrat A., Prendergast G.C., Pavlath G.K. (2013) The N-BAR domain protein, Bin3, regulates Rac1- and Cdc42-dependent processes in myogenesis. Dev. Biol., 382, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rankin E.B., Higgins D.F., Walisser J.A., Johnson R.S., Bradfield C.A., Haase V.H. (2005) Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol. Cell Biol., 25, 3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Alvarez-Buylla A., Ling C.Y., Kirn J.R. (1990) Cresyl violet: a red fluorescent Nissl stain. J. Neurosci. Methods, 33, 129–133. [DOI] [PubMed] [Google Scholar]
- 90. Makinson C.D., Dutt K., Lin F., Papale L.A., Shankar A., Barela A.J., Liu R., Goldin A.L., Escayg A. (2016) An Scn1a epilepsy mutation in Scn8a alters seizure susceptibility and behavior. Exp. Neurol., 275 Pt 1, 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Crawley J.N. (2000) What’s Wrong With My Mouse: Behavioral Phenotyping of Transgenic and Knockout Mice. John Wiley & Sons, Inc, New York. [Google Scholar]
- 92. Prusky G.T., Alam N.M., Beekman S., Douglas R.M. (2004) Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest. Ophthalmol. Vis. Sci., 45, 4611–4616. [DOI] [PubMed] [Google Scholar]
- 93. Aung M.H., Kim M.K., Olson D.E., Thule P.M., Pardue M.T. (2013) Early visual deficits in streptozotocin-induced diabetic long evans rats. Invest. Ophthalmol. Vis. Sci., 54, 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Aung M.H., Park H.N., Han M.K., Obertone T.S., Abey J., Aseem F., Thule P.M., Iuvone P.M., Pardue M.T. (2014) Dopamine deficiency contributes to early visual dysfunction in a rodent model of type 1 diabetes. J. Neurosci., 34, 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chekanova J.A., Shaw R.J., Belostotsky D.A. (2001) Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol., 11, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 96. Apponi L.H., Leung S.W., Williams K.R., Valentini S.R., Corbett A.H., Pavlath G.K. (2010) Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet., 19, 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M.. et al. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity, 32, 279–289. [DOI] [PubMed] [Google Scholar]
- 98. Zambon A.C., Gaj S., Ho I., Hanspers K., Vranizan K., Evelo C.T., Conklin B.R., Pico A.R., Salomonis N. (2012) GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics, 28, 2209–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Csardi G. N.T. (2006) The igraph software package for complex network research. InterJournal Complex Systems, 1695. [Google Scholar]
- 100. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 101. Cheever A., Blackwell E., Ceman S. (2010) Fragile X protein family member FXR1P is regulated by microRNAs. rna, 16, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.