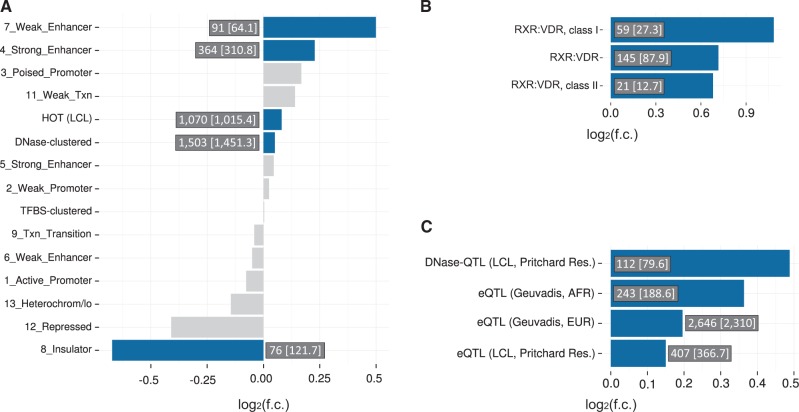
Figure 3.
Genomic association testing of VDR-BVs with functional annotation. VDR-BVs are enriched at enhancers, in RXR::VDR DR3-type motifs, and in LCL eQTLs. (A) Enrichment of VDR-BVs within ENCODE chromatin state segmentation tracks for LCL NA12878, TF-dense regions (HOT (17) and ENCODE clustered TF binding sites) and DNase hypersensitive areas (ENCODE). (B) Enrichment of VDR-BVs in VDR consensus motif intervals present in VDR CPo3 binding regions. (C) Enrichment of VDR-BVs at DNase-QTLs and eQTLs from the Pritchard resource and CEU/YRI LCL eQTLs from the GEUVADIS resource. Significant enrichments are indicated using blue histogram bars (f.c. = fold change of observed versus expected overlaps; FDR q < 0.1; numbers in a box close to each bar indicate ‘observed [expected]’ overlap counts; light grey bars indicate lack of significance). For A, enrichments shown are above-and-beyond the previously observed (Figure 1E) enrichments of VDR CPo3 peaks in the same functional annotation classes (relative to a background of 20,330 1000 Genomes SNPs in CPo3 VDR binding regions). For B, enrichments are relative to a background of 114,155 1000 Genomes variants lying under VDR ChIP-exo read pileups (≥ 5 reads) which had been tested as potential VDR binding affinity modifiers. This background was further corrected for the analyses in C, were only LD-independent foreground VDR-BVs were tested and 10,000 DAF-matched random background sets were extracted with replacement from the main set of 114,155 background variants.