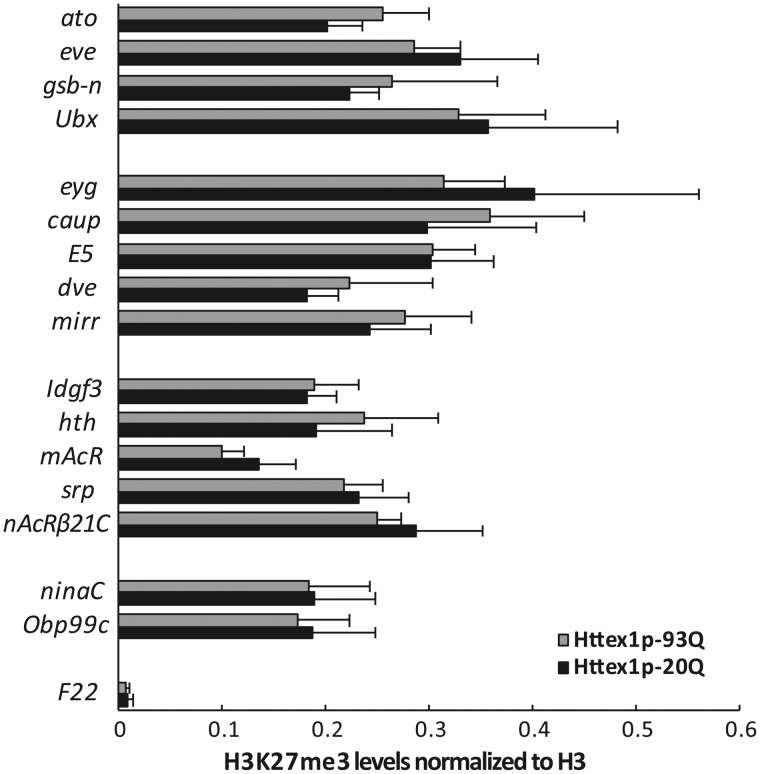
Figure 5.
H3K27me3 occupancy is not altered on candidate genes in mutant Htt expressing flies. The level of H3K27me3 modification on genes with H3K27me3 enriched regions was determined by ChIP-qPCR in head samples of Httex1p-Q93 or Httex1p-Q20 (control) expressing flies. The tested gene set included genes not expressed in neurons (ato, eve, gsb-n, Ubx), and ones with low (eyg, caup, E5, dve, mirr), moderate (Idgf3, hth, mAcR, srp, nAcR-beta21C) or high (ninaC, Obp99c) expression levels, and a negative control region (F22) was also included. Significant differences were observed in ChIP signal intensity between the tested loci (P = 1.04 × 10−5, two-way ANOVA) but not in H3K27me3 levels between Httex1p-Q93 and Httex1p-Q20 expressing flies (P = 0.961, two-way ANOVA). Bars show the ratio of average ChIP-qPCR signals for H3K27me3 normalized to total input control and that for histone H3. Error bars represent standard error of mean (n = 3).