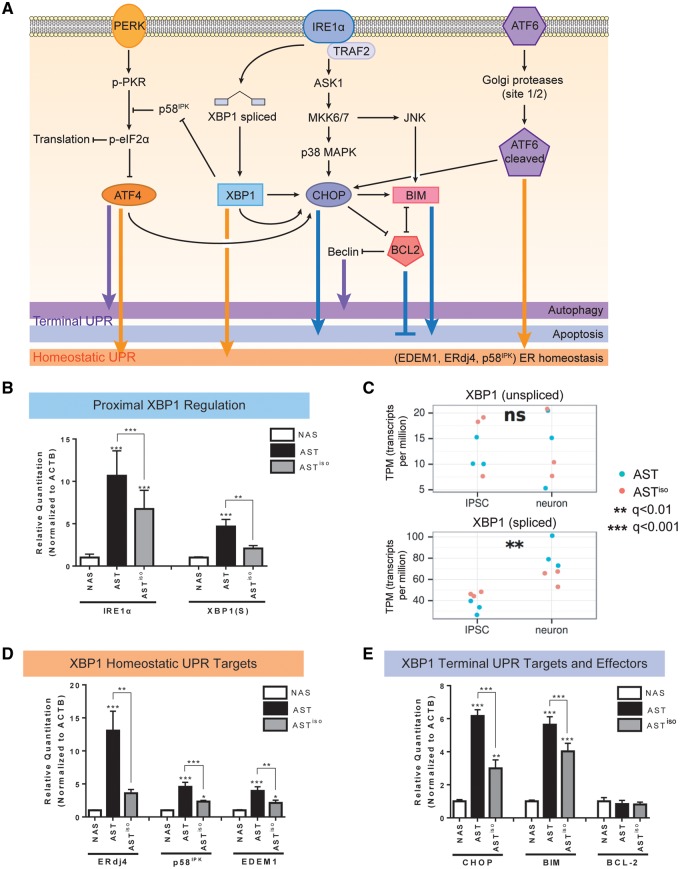
Figure 3.
Transcript and protein expression validation of IRE1α/XBP1-induced UPR activation in SNCA triplication iPSC-derived neurons. (A) Schematic of signaling pathways downstream of the UPR. PERK, IRE1α and ATF6 branches are indicated, and the effects of downstream effectors on homeostatic, autophagic and apoptotic outcomes are indicated. (B, D, E) Validation of mRNA expression level differences in IRE1α/XBP1 pathway components by qRT-PCR. Levels of IRE1α and spliced XBP1 [XBP1(S)] (B), the homeostatic UPR targets ERdj4, p58IPK and EDEM1 (D), and the terminal UPR targets CHOP, BIM and BCL-2 (E) at the mRNA level were evaluated by qRT-PCR in NAS, AST and ASTiso iPSC-derived neurons. Data are represented as mean ± SEM of biological triplicates. (C) Gene expression from RNAseq data (in transcripts per million, TPM) of unspliced and spliced XBP1 transcripts across differentiation stage (iPSC or neuron) and genotype (blue = AST, red = ASTiso). *P ≤ 0.5, ** P ≤ 0.01, *** P ≤ 0.001, ns = not significant. See also Supplementary Material, Figs S5–S7 and Table S4.