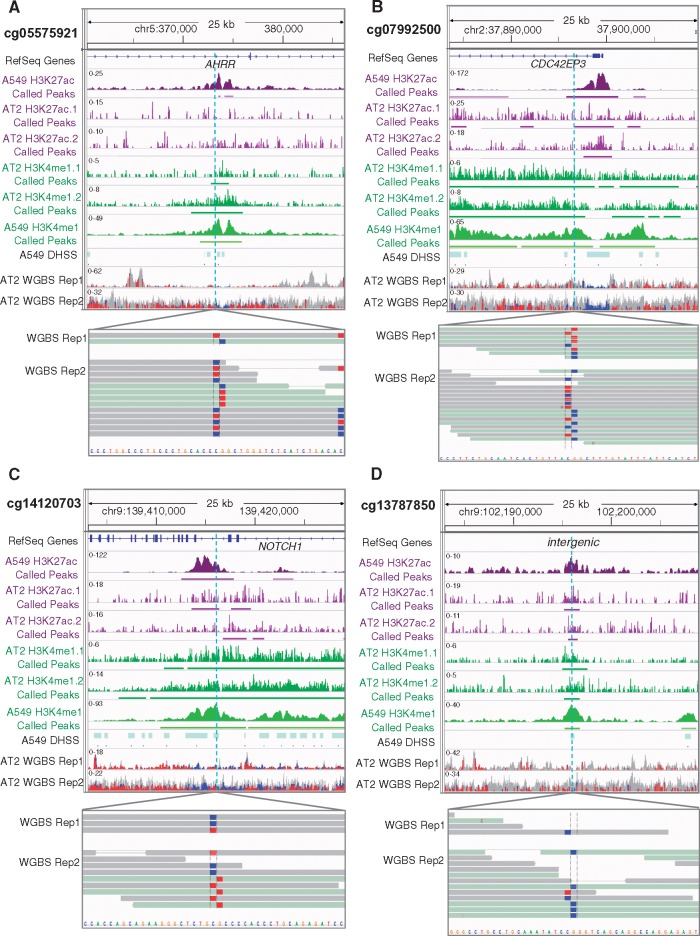
Figure 1.
Four CpGs hypomethylated in smoker NTL are adjacent to unmethylated loci in AT2 cells and marked by regulatory histone marks in AT2 and A549 cells. Displayed from top to bottom for each CpG: the genomic location with RefSeq genes (if present; hg19 coordinates); H3K27ac (purple) & H3K4me1 (green) profiles in A549 cells and in two biological replicates of AT2 with called peaks underlined; DNase hypersensitive sites in A549; and two biological replicates of whole genome bisulfite sequencing (WGBS) with the CpG of interest magnified (blue= unmethylated; red = methylated; gray = no CpG present). A549 data were downloaded from ENCODE (45). The sequencing depth of AT2 cells is less than that of the A549 cell line owing to the difficulty in obtaining large numbers of primary human cells.