Abstract
Although mitochondrial genomes (mtDNA) accumulate elevated levels of mutations in cancer cells, the origin and functional impact of these mutations remain controversial. Here, we queried whole-genome sequence data from 1,916 patients across 24 cancer types to characterize patterns of mtDNA mutations and elucidate the selective constraints driving their fate. Given that mitochondrial genomes are polyploid, cells with advantageous levels of mtDNA mutations can be selected for depending on their cellular environment. Therefore, we tracked changes in per-cell abundances of mtDNA mutations from normal to tumor cells in the same patient. Tumor mitochondrial genomes show distinct mutational patterns and are disproportionately enriched for protein-altering changes. Moreover, protein-altering mtDNA variants that are initially present at low frequencies in normal cells preferentially expand in the altered tumor environment, suggesting selective advantage. We also perform these analyses with attention to the cancer’s tissue of origin, which revealed tissue-specific differences in selective signals. The mitochondrial genomes in renal chromophobe and thyroid cancers show particularly strong signals of positive selection, indicated by higher proportions and per-cell abundances of truncating variants. Dramatic tumor- and tissue-specific variations in selective pressures suggest that cancer cells with advantageous levels of damaged mitochondrial genomes will selectively proliferate to facilitate the tumorigenic process.
Introduction
Having originated from single-celled organisms, mitochondria carry their own genomes and are widely regarded as the energy powerhouses of the cell. Over time, much of the mitochondrial genetic content has migrated to the cell’s nucleus. In humans, mitochondrial genomes have retained some 16.5 kilobase pairs of circular, double-stranded DNA harboring 37 genes. Thirteen of these genes are protein-coding and form key components of four of the five energy-generating complexes collectively known as the electron transport chain (ETC). Mutations in mtDNA have been shown to impair energy production, generate elevated levels of damaging by-products known as reactive oxygen species (ROS) (1–3), and can cause severe metabolic disorders (4). MtDNA becomes damaged at a rate an order of magnitude higher than the nuclear genome (5), making it an appealing candidate for involvement in cancer progression and for understanding patterns of Darwinian selection in normal (6) and tumor cell environments alike. Nonetheless, there have been conflicting reports regarding the function of mitochondria and their genomes in the tumorigenic process (7–13).
Multiple studies have demonstrated strong purifying selection acting on mtDNA in the human germline (6,14–16). These observations have been recapitulated experimentally, with both mouse (17) and Drosophila (18,19) experiments showing strong selection against transmission of deleterious mtDNA variants to offspring. However, the question of whether there is selection in somatic cells remains unsettled. Studies of human brain (20) and colon epithelial tissues (21) showed no evidence of selection in the soma. In contrast, others report signals of purifying selection against deleterious mtDNA mutations in blood (22), and of positive selection for somatic mutations, particularly in liver (23). Non-random appearance of tissue-specific mutations (23–25) supports the notion that the presence and degree of selective pressures on human mtDNA variants are strongly dependent on tissue type. This dependence could conceivably extend to tumor cells, with selective conditions varying with tumor type. The goal of our study is to investigate, in a tumor type-specific manner, the signals of selection present in the mtDNA mutational landscape of human cancer.
A small number of recent studies have examined pan-cancer mtDNA mutational (13,26) and copy-number (27) patterns. However, these studies focused primarily on tumor-specific aberrations. Mutations already present in normal tissue—whether inherited or acquired—may be subject to selective pressures in the neoplastic cell. Since multiple mtDNA haplotypes may be present in the same cell or tissue—a condition known as heteroplasmy—the tumor has the ability to respond to altered selective pressures through dramatic changes in allelic frequencies. Tracking mtDNA variants from normal to tumor tissues allows assessment of selective pressures acting on individual mtDNA copies and their overall per-cell abundances. Performing these analyses in the context of the cancer’s tissue of origin reveals differences in selective signals that would otherwise be obscured.
Here we analyzed mitochondrial sequences generated by the Cancer Genome Atlas (TCGA) (28) using deep next-generation sequencing (NGS) data from 1,916 patients spanning 24 cancer types. In order to understand the selective pressures governing mitochondrial genome integrity, we identified patterns of mtDNA variants in both tumor (derived from primary, recurrent, and/or metastatic tumors) and matched normal samples (derived from blood and/or adjacent tissues) (Fig. 1A;Supplementary Material, Tables S1–S3). We queried for normal-cell variants whose heteroplasmy levels shift in the tumor, in addition to tumor-specific somatic mutations absent from normal-cell data. Our results suggest that tumor cells generally show neutral evolution of their mitochondrial genomes as compared to the stringent negative selection found in the human germline. Moreover, these relaxed selective constraints allow for dramatic expansions of frameshift and nonsense mtDNA alleles in tumor cells, particularly in endocrine and rare kidney cancers and may define tumor types that are driven by mtDNA variants. Our results also show positive selection acting via preferential increase in allelic frequencies of non-synonymous mtDNA variants in the tumor.
Figure 1.
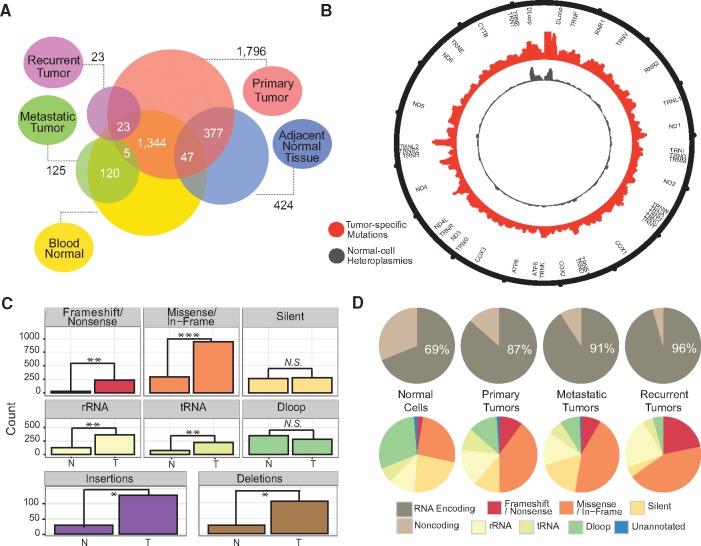
Tumor mitochondrial genomes show distinct mutational features. (A) Venn diagram of all TCGA samples and sample types under consideration. Numbers within the circle intersections indicate numbers of patients with the corresponding sample types available. (B) Circos plot representing mtDNA variant distribution across the mitochondrial genome plotted using 50 base pair sliding windows. (C) Counts of tumor-specific somatic mutations (T) and normal cell heteroplasmies (N) stratified by genome annotation (* signifies P < 1 × 10−15, **P < 1 × 10−30, ***P < 1 × 10−45, and N.S. = not significant). (D) RNA encoding vs. non-coding (TOP; omnibus P = 5.40 × 10−37) and variant annotation (BOTTOM) proportions for various sample types.
Results
Tumor mitochondria have distinct genomic features
We designed and implemented a pipeline tailored to extract mitochondrial reads and call sequence variants (29). We restricted our analysis to patients with whole-genome sequencing (WGS) data from both tumor and matched normal DNA to ensure sufficient read depth for accurate variant calling (Supplementary Material, Figs S1–S3). As quality control measures, we removed artifacts (Supplementary Material, Fig. S4), mismatched samples, and DNA contamination (Supplementary Material, Note S1). Distinguishing between truly tumor-specific somatic mutations and those already present in normal cells is not straightforward, nor is the distinction between germline and somatically-acquired variants in the normal cell. Variants observed in normal cells may have had selection act upon them over the course of multiple generations, including repeated maternal germline bottlenecks, or may have been acquired sporadically and expanded either through random drift or selective pressures (Supplementary Material, Note S2) (30). Similarly, tumor-specific mutations either were already present within the initiating malignant cell or arose subsequently. An mtDNA variant harbored within the genome of the tumor’s founder cell can be carried forward through clonal expansion. It follows that such variants may be below any assay’s level of detection in the normal-tissue biopsy, but may become apparent in the tumor. In either case, these mutations have been subjected to any altered selective pressures present since neoplastic initiation.
In our cohort, we observed a total of 2,350 tumor-specific somatic mtDNA mutations (across 1,231 individuals, 64.3%) (Supplementary Materials, Fig. S5 and Table S4) and 1,154 normal-cell heteroplasmies (across 760 individuals, 39.7%) (Supplementary Material, Table S4). Tumor mitochondrial genomes carry distinct mutational signatures (Supplementary Materials, Figs S6 and S7), and show changes in their distribution of mutations (Fig. 1B). While normal-cell heteroplasmies tend to cluster within the non-coding D-loop, tumor-specific somatic mutations are more evenly dispersed across both coding and non-coding regions (Fig. 1B). We see higher numbers of tRNA, rRNA, and mRNA aberrations in tumors than in normal cells, whereas we see little difference in silent and non-coding (D-Loop and unannotated) variant levels (Fig. 1C). The proportion of mtDNA variants within RNA-encoding regions (as opposed to non-coding regions) is larger for tumor-specific mtDNA mutations than normal-cell heteroplasmies (Fisher’s exact P = 2.17 × 10−33). This difference could be the result of altered selective pressures in the tumor. Alternatively, it could instead be the result of the RNA variants being more disruptive and therefore present at lower levels (below the threshold of detection) in the normal cell—either owing to purifying selection in the germline or insufficient time to drift to higher levels after somatic acquisition—but clonally expanding in the tumor with the initiating cell. Interestingly, however, metastatic and recurrent tumors harbor an even larger proportion of RNA variants vs. D-loop/unannotated variants than do primary tumors (P = 0.021) (Fig. 1D). Although the numbers of metastases and recurrences with available TCGA WGS data is small (81 metastases and 16 recurrences), this suggests the possibility that the clone seeding the distant metastasis, or resistant to standard treatment, is more likely to harbor RNA-encoding mutations. Tumor-specific somatic mutations also have greater representation of insertions and deletions (indels) (9.2% of variants) (Fig. 1C) compared to normal-cell heteroplasmies (2.95% of variants) (Supplementary Material, Table S5).
Variants were distributed across all complexes of the ETC (Supplementary Materials, Figs S8–S11, Table S6, and Note S3), but there were several recurrent and mutually exclusive tumor-specific mutations within genes encoding for the NADH dehydrogenase complex (Complex I), the initial electron acceptor along the ETC (Supplementary Materials, Figs S8D, S12, S13, Table S6, Note S4) (although it should be noted that Complex I has the largest total coding region among the complexes, and may therefore be more likely to acquire mutations randomly). Such recurrences have been noted in separate human populations over recent evolutionary time (31), and a similar phenomenon may be at work in the tumor. Tumor types with typically high nuclear somatic mutational burden are not necessarily those with higher mtDNA mutational burden. However, we found statistically significant relationships between nuclear and mitochondrial somatic events (Supplementary Materials, Figs S14 and S15, Note S5) including marked discordance between protein-altering mutations in ETC genes encoded in the two cellular compartments (Supplementary Materials, Fig. S16 and Note S6).
Tissue-specific variation in selective pressures on tumor mitochondrial genomes
Higher proportions of protein-altering mtDNA mutations in tumor cells indicates increased tolerance for disruptive mtDNA variants. To investigate this formally, we assessed selection on protein-coding genes by comparing the rate of non-synonymous mutations to that of synonymous mutations (32). A higher non-synonymous rate than what would be expected by chance would indicate positive selection, while a lower-than-expected rate would indicate negative selection. To this end, we simulated the null hypothesis of no selection (random mutation) by performing a permutation analysis while controlling for mutational biases due to base content (i.e. the mutated nucleotide residue and its two flanking bases) (Supplementary Material, Fig. S17) (13). Comparing observed ratios of non-synonymous to synonymous mutation rates (dN/dS) to the permutation-derived null distribution, normal-cell heteroplasmies had a far lower than expected dN/dS (P < 10−3), showing strong negative selection against protein-altering variants. In contrast, tumor-specific somatic mtDNA mutations as a whole showed no evidence for positive or negative selection by this measure (P = 0.228). P-values for somatic mutations by gene and by tumor type are distributed roughly uniformly across the unit interval, indicating largely neutral tolerance for non-synonymous tumor-specific somatic mtDNA mutations (Fig. 2A and B; see Supplementary Material, Table S1 for tumor type abbreviation key).
Figure 2.
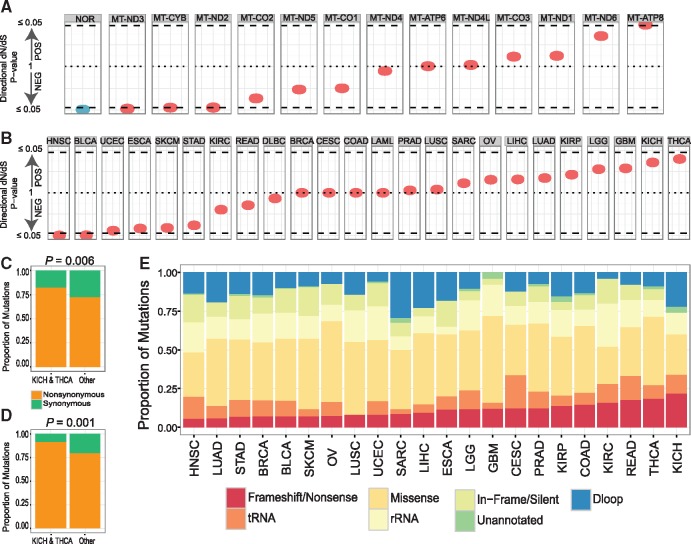
Tissue-specific signals of selection on tumor mitochondrial genomes. (A) Nominal P-values derived from dN/dS permutation analysis to indicate direction and statistical significance of selection on protein-altering variants in normal (NOR, blue), and tumor cells (red) across mitochondrial genes and (B) across individual cancer types. (C) Proportions of synonymous and nonsynonymous mtDNA mutations including both tumor-specific somatic variants and those already present in the normal cell and (D) tumor-specific somatic variants only. (E) Proportions of mtDNA variants of various categories, stratified by tumor type.
Among all tumor types KICH and THCA showed dN/dS results most consistent with positive selection (Fig. 2B). For KICH and THCA patients, 79.6% of coding SNVs in their tumor cells were non-synonymous, as compared to 70.3% or all other tumor types (P = 0.006) (Fig. 2C). For tumor-specific somatic mutations, the difference between these ratios was even more prominent, at 88.9% for KICH/THCA patients and 77.6% for other tumor types (P = 0.001) (Fig. 2D). Therefore, these two tumor types have a higher rate of protein-altering mutations compared to other tumor types, when background mutation rate is controlled for. Additionally, KICH and THCA showed the highest overall proportions of frameshift/nonsense mutations (KICH proportion vs. other tumor types P = 8.774 × 10−3; THCA P = 4.236 × 10−4) (Fig. 2E). KICH patients showed particular enrichment for transversions (∼19.4%, P = 1.2 × 10−3) (Supplementary Material, Fig. S18), which are more likely to cause amino acid changes. Interestingly, previous reports show that KICH cells have significant enrichment of metabolic pathways related to mitochondrial energy production (33). These results demonstrate that, although most tumor types exhibit neutral evolution of their mitochondrial genomes, KICH and THCA cells show evidence for positive selection acting on their mtDNA.
Low-level non-synonymous mtDNA alleles in normal cells clonally expand in tumors
Each cell can carry hundreds to thousands of copies of the mitochondrial genome. The impact of mtDNA variants that are deleterious to the cell can therefore be mitigated if they remain at a low allelic frequency. This is not achievable in the diploid nuclear genome since variants must be present in at least one of two copies. Some low-level mtDNA variants may dramatically increase in frequency if they confer a selective advantage in the altered tumor environment, or may increase or decrease through random drift if they are selectively neutral in that environment. Equipped with allelic frequency data in tumor and normal tissue from the same patients (Supplementary Material, Note S7), we next sought to investigate the dynamics of heteroplasmy from normal to tumor tissues, querying for signals of selection. In normal-cell heteroplasmies, we deem one allele as ancestral (inferred by comparing human mtDNA sequence with that of non-human primates (22)) and the other allele as derived. Heteroplasmy levels are then specified by their derived allele frequencies (DAFs).
We stratified DAFs of heteroplasmic variants by effect on protein—synonymous substitution, missense mutation (in-frame indels and missense substitutions) or truncating mutation (frameshift indels and nonsense substitutions). In normal cells, missense mutations are at significantly lower allelic frequencies than synonymous substitutions, but are at much higher abundances than the more disruptive truncating variants (Fig. 3A). In the tumor, these same variants showed no difference in DAF among the three classes (Fig. 3B). Tumors also demonstrated a much higher tolerance for disruptive somatic mtDNA mutations, with no statistical association between their mutant allelic frequencies (MuAF) and variant class (Fig. 3C). Similarly, tRNA-coding regions showed a higher average tumor MuAF than normal cell DAF (Supplementary Material, Note S8).
Figure 3.
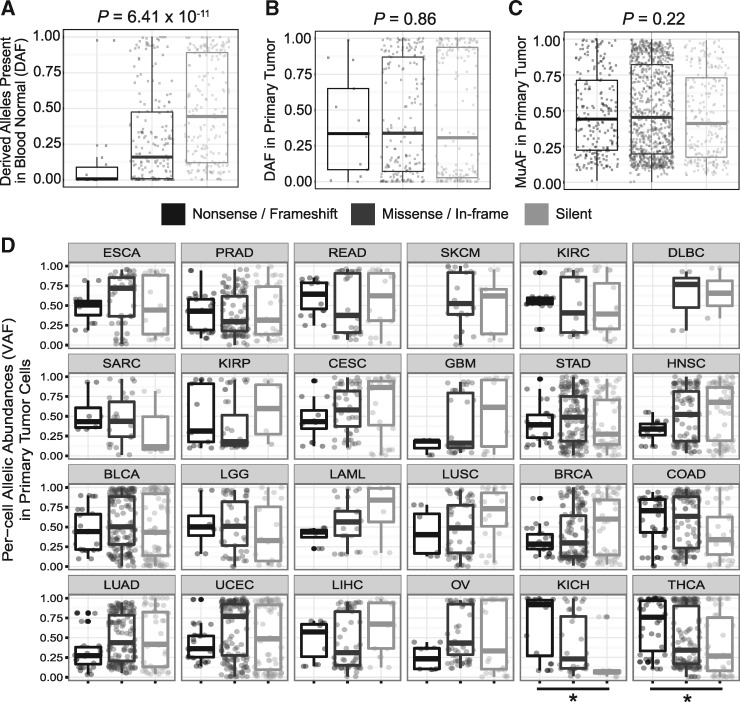
Tissue-specific signals of relaxed and positive selection on mtDNA allelic abundances in tumor cells. Stratified by their effects on the protein: nonsense/frameshift, missense/in-frame, and silent, distributions of derived allele frequencies (DAFs) of (A) variants present in blood normal (Kruskal-Wallis, P = 6.41 × 10−11) and (B) primary tumor variants which were also present in the normal (P = 0.86). (C) Distributions of mutant allele frequencies (MuAFs) of tumor-specific somatic mutations present only in primary cells (P = 0.22). (D) Per-cell allelic abundances of all variants present in primary tumor cells stratified by cancer type, * indicates Jonckheere’s trend test P-values < 1 × 10−2 (P = 1.6 × 10−3 for KICH and P = 1.55 × 10−4 for THCA).
Previous studies have shown that specific normal tissue types have consistent somatic mutational patterns across individuals (23–25). To investigate this tissue-specificity in the context of tumors, we queried coding variant allele frequencies (VAFs) in tumor cells for both somatic and derived alleles as delineated by tumor type. While most tumor types corroborate the finding that there are no differences in allelic frequencies between the three classes of mutations, we found that kidney chromophobe (KICH) and thyroid (THCA) cancers have VAFs that correlate positively with the severity of mutation class (Fig. 3D, Jonckheere trend test P = 1.6 × 10−3 for KICH and P = 1.55 × 10−4 for THCA). This may be indicative of positive selection in KICH and THCA, given that the cohort as a whole seems to demonstrate overall neutral evolution.
Preferential expansions of low-level non-synonymous alleles suggests positive selection for mtDNA variants
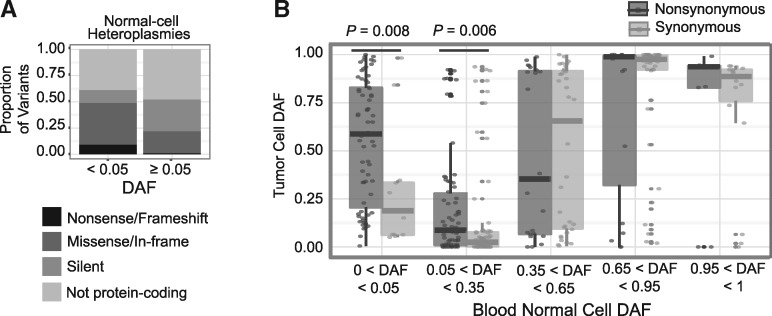
We next restricted attention to alleles that were present at low levels in the normal cell. The rationale here is that many such alleles are at low abundance due to their deleterious effects on the cell, but may be subject to different selective pressures in the dysregulated tumor. As expected, alleles below a 5% level in the normal cell were highly enriched for deleterious variants as compared to those above a 5% level (non-synonymous enrichment P = 3.8 × 10−10; frameshift/nonsense enrichment P = 8.3 × 10−5; Fig. 4A). Non-synonymous substitutions below 5% in the normal cell expand to a median allelic frequency of 58.8% in the tumor, as compared to 18.8% for synonymous substitutions (P = 0.008), suggesting positive selection in the tumor (Fig. 4B). Furthermore, all three frameshift insertions that were present at these low frequencies in the normal cell expanded substantially, to 31, 43, and 85% allelic frequencies in the tumor.
Figure 4.
Disruptive low-level mtDNA variants preferentially expand in the tumor. (A) Proportions of each variant type are shown for all variants with DAF < 5% and ≥ 5%. (B) Tumor cell DAF of nonsynonymous vs. synonymous variants, stratified by normal cell DAF binned into five incremental categories.
The trend of preferential expansion of non-synonymous substitutions also holds for variants present at intermediately low frequencies (5–35%) in the normal cell, with tumor DAFs significantly larger for non-synonymous variants than synonymous (P = 0.006; Fig. 4B). For variants with higher normal-cell DAFs (>35%), however, this trend does not hold. Altogether, these results suggest positive selection in tumor cells for disruptive mtDNA variants present at low levels in normal cells.
Individuals with deleterious alleles that dramatically expand in the tumor are dominated by thyroid and kidney cancer patients
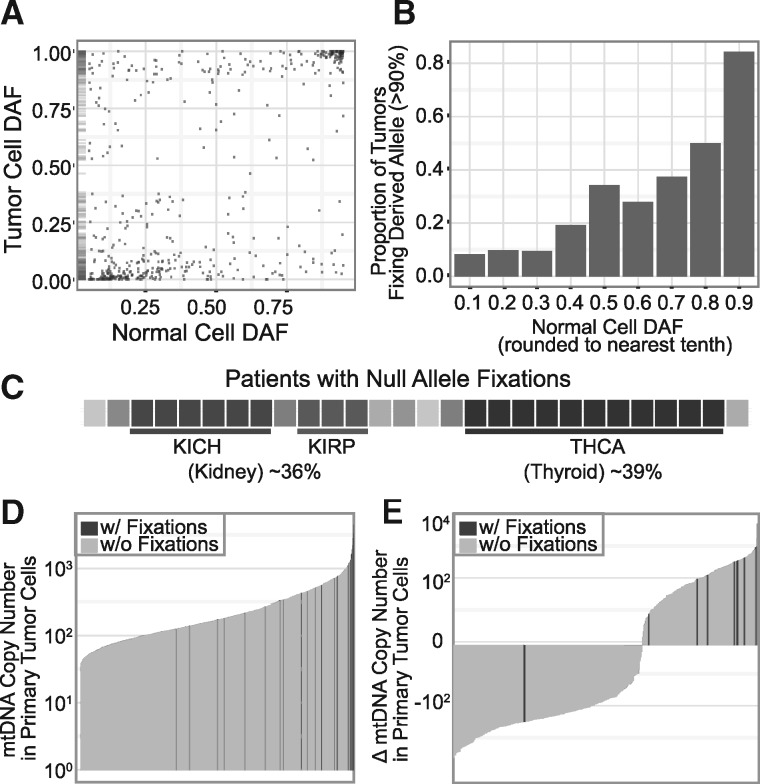
Overall, normal-cell heteroplasmies tend toward homoplasmy in the tumor (Fig. 5A). Much of this effect is likely due to the effective bottleneck formed by the tumor’s expansion from a single initiating cell whose composition is more homoplasmic than the bulk normal-tissue sample. Indeed, the likelihood of a derived allele approaching fixation in the tumor is closely tied to its frequency in the normal cell (Fig. 5B), as would be expected under a model of random drift (34). However, as noted above, there are alleles that dramatically diverge from this tendency (e.g. upper left corner of Fig. 5A). Even null alleles (nonsense and frameshift) present at low frequency in the normal cell can become dominant in the tumor. We therefore next undertook a closer examination of patients harboring truncating or frameshift alleles that were non-existent or at very low frequency (< 5%) in the normal cell but rose to near-fixation (> 90%) in the tumor. We hypothesized that these may be the patients for which mtDNA mutations play a greater functional role in tumorigenesis and progression. We observe this pattern of ‘null allele fixation’ in 28 patients (Table 1). Despite making up fewer than 7% of all patients, THCA comprises 39% of all cases of null allele fixation (P = 3.8 × 10−10). The three TCGA kidney cancer subtypes collectively were also overrepresented, comprising 36% of null allele fixation cases (P = 1.5 × 10−9) despite making up only 5.9% of tumors in the patient population. We observed particular enrichment in KICH and kidney renal papillary cell carcinoma (P = 2.9 × 10−9 and 4.9 × 10−3) (Fig. 5C).
Figure 5.
Null alleles present at extremely low levels in the normal rise to near-fixation in the tumor. (A) DAF in tumor cells plotted versus normal cell (starting) DAF; rug plot on the left shows density of points along the y-axis. (B) Proportion of variants that achieve fixation (defined as DAF > 90%) stratified by normal cell (starting) DAF. (C) Distribution of cancer types among patients with null allele fixations, each shade represents a unique tumor type. With null allele fixators indicated in black, barplots of (D) mtDNA copy numbers in all primary tumor samples and (E) change in mtDNA copy number in primary tumor cells from matched adjacent normal tissue cells.
Table 1.
Null allele fixations
| Patient | Position | Ancestral Allele | Alternate Allele | Normal Alternate Allele Frequency | Tumor Alternate Allele Frequency | Gene | Alternate Allele Effect |
|---|---|---|---|---|---|---|---|
| CESC_A3LA | 6691 | GA | G | 0.000 | 0.947 | COX1 | frameshift_variant |
| COAD_3666 | 12417 | CA | C | 0.000 | 0.940 | ND5 | frameshift_variant |
| KICH_8343 | 3565 | A | AC | 0.018 | 0.971 | ND1 | frameshift_variant |
| KICH_8344 | 13127 | AC | A | 0.000 | 0.984 | ND5 | frameshift_variant |
| KICH_8419 | 12417 | C | CA | 0.000 | 0.954 | ND5 | frameshift_variant |
| KICH_8430 | 13206 | CTG | C | 0.000 | 0.955 | ND5 | frameshift_variant |
| KICH_8439 | 14619 | A | AC | 0.000 | 0.997 | ND6 | frameshift_variant |
| KICH_8477 | 10806 | G | A | 0.000 | 0.986 | ND4 | stop_gained |
| KIRC_5453 | 12384 | T | TC | 0.000 | 0.917 | ND5 | frameshift_variant |
| KIRP_A40X | 5490 | CCT | C | 0.000 | 0.911 | ND2 | frameshift_variant |
| KIRP_A48D | 14503 | TA | T | 0.000 | 0.960 | ND6 | frameshift_variant |
| KIRP_A4EM | 12417 | CA | C | 0.000 | 0.966 | ND5 | frameshift_variant |
| LGG_7675 | 11888 | G | A | 0.000 | 0.964 | ND4 | stop_gained |
| PRAD_5765 | 3408 | C | CT | 0.000 | 0.943 | ND1 | frameshift_variant +stop_gained |
| SARC_A3KA | 10207 | CT | C | 0.000 | 0.931 | ND3 | frameshift_variant |
| STAD_6875 | 11403 | G | A | 0.008 | 0.971 | ND4 | stop_gained |
| THCA_A13R | 11390 | G | A | 0.000 | 0.977 | ND4 | stop_gained |
| THCA_A13S | 12417 | C | CA | 0.000 | 0.991 | ND5 | frameshift_variant |
| THCA_A1CW | 6753 | G | A | 0.000 | 0.932 | COX1 | stop_gained |
| THCA_A1YA | 11034 | A | AT | 0.000 | 0.998 | ND4 | frameshift_variant |
| THCA_A1YB | 12417 | CA | C | 0.000 | 0.989 | ND5 | frameshift_variant |
| THCA_A28R | 9502 | G | A | 0.000 | 0.975 | COX3 | stop_gained |
| THCA_A2OW | 4720 | G | A | 0.001 | 0.987 | ND2 | stop_gained |
| THCA_A3I5 | 9241 | A | G | 0.000 | 0.963 | COX3 | stop_gained |
| THCA_A3MY | 11866 | A | AC | 0.004 | 0.913 | ND4 | frameshift_variant |
| THCA_A3O9 | 11403 | G | A | 0.000 | 0.989 | ND4 | stop_gained |
| THCA_A3RA | 8618 | T | TC | 0.000 | 1.000 | ATP6 | frameshift_variant |
| UCEC_A1GW | 6579 | G | A | 0.000 | 0.983 | COX1 | stop_gained |
All reported null allele fixations found in 28 patients in our cohort, their mtDNA positions, allelic frequencies in the normal and tumor cells, and respective mutational effects.
Next, we analyzed the dynamics of null allele fixations with respect to changes in the number of mitochondrial genome copies per cell (Supplementary Materials, Figs S19 and S20); mtDNA copy number correlates with other molecular (Supplementary Material, Fig. S21) (27) and clinical features (Supplementary Material, Fig. S22). The 28 patients with null allele fixations tend to have much higher mtDNA copy numbers in their primary tumor cells (median copy number 729, vs. 165 for other patients, P = 2.7 × 10−10) (Fig. 5D). This difference remained highly significant even after controlling for tumor type. Nine of the patients with these fixations had data available from adjacent normal tissue. Shifts in copy number from adjacent normal tissue to the tumor are also markedly larger in patients with null allele fixations, gaining a median 400 mtDNA copies as compared to an average loss of 59 copies for the other patients. All but one (89%) gained copies, compared to only 37% of patients without null allele fixations (P = 0.0047) (Fig. 5E). Using blood as a normal comparator yields even more dramatic shifts in copy number (median gain 858 copies vs. 43 in patients with and without null allele fixations, respectively, P = 5.3 × 10−8). We also observed statistically significant associations between fixation status and expression of genes involved in programmed cell death (apoptosis) which is a key cellular housekeeping function of healthy mitochondria (Supplementary Material, Note S9).
Discussion
Mitochondria have retained their own genomes in nearly all eukaryotic cells, although the significance of their retention is unclear (35–37). The role of mitochondrial genomes in human cancer cells also remains controversial. To date, large-scale cancer mutation studies have focused almost exclusively on the nuclear genome with only a few querying mtDNA (13,26,27,38). Unlike the diploid nuclear genome, copies of the mitochondrial genome range from hundreds to thousands per cell and vary both spatially and temporally, allowing for fluctuations in cellular levels of mtDNA variants. Therefore, mitochondrial genomes provide a sensitive readout for altered selective pressures in the cell and can facilitate our understanding of the role of mitochondria in cancer. Here we report novel insights into selective pressures acting on tumor mitochondrial genomes and the resulting differences in the levels of damaged mtDNA copies between normal and tumor cells.
The work presented here bolsters the hypothesis that mtDNA variation contributes to human cancer. We show that tumor cells are highly tolerant of protein-altering mutations. This tolerance manifests in high numbers of mutations, a large proportion of non-synonymous coding variants, as well as an elevated per-cell allele frequency of these non-synonymous variants. The overall relaxation of selective constraints in tumor cells provides opportunities for expansions of certain mutated mtDNA copies. Indeed, tumor cells show positive selection via preferential expansion of non-synonymous mutations present at low levels in normal cell. We also observe frameshift and nonsense variants—which likely abrogate protein function—that are absent or near-absent in the normal cell but rise to dominance in the tumor. Intriguingly, these fixations are generally accompanied by dramatic increases in total mtDNA copy number which indicates that mitochondrial genomes harboring these null alleles are replicated at an unusually high rate in the tumor and may have a replicative advantage (18,19). Tumor cells therefore display an overall increased tolerance and/or some selective preference for damaged mitochondrial genomes.
Although most tumor cells show neutral evolution of their mitochondrial genomes, we have multiple lines of evidence suggesting positive selection on mtDNA in kidney chromophobe and thyroid cancers in particular. Patients with these tumor types harbor significantly higher proportions and per-cell abundances of nonsynonymous over synonymous changes than do other tumor types. Interestingly, a recently published study of mtRNA expression in TCGA found that KICH and THCA are the only tumor types (of 13) that show recurrent increase in mitochondrial gene expression in tumors (as compared to matched normal tissue) (39). Previous studies report that kidney chromophobe and thyroid cells have specialized mitochondria; KICH cells have significant enrichment of metabolic pathways related to mitochondrial energy production (33) and thyroid cells have increased dependence on mitochondrial oxidative phosphorylation (40). KICH and THCA are also generally less malignant and slow-growing (41), suggesting that extremely damaged mitochondria may not be ideal for tumor progression and that tumor cells may instead benefit from having moderate levels of damage to their mtDNA (42). The positive selective signals characterize KICH and THCA as exceptions to the rule that tumor mitochondria undergo neutral evolution of their genomes.
Two recent studies also examined patterns of selection in tumor mtDNA using TCGA data. Ju and colleagues (13) analyzed 1675 tumors (a minority of which were from TCGA), and concluded that selection was largely neutral on the mitochondrial genome in cancer cells. Broadly, their conclusions do not differ substantially from ours. Indeed, we find that the negative selection present in the germline is largely relaxed in cancer, though we conclude that positive selection does act on a subset of tumors. The partial discrepancy in conclusions between our study and the Ju et al. study is likely the result of differences in the compositions of tumor types. A substantial portion of the positive selection signal in our data comes from THCA and KICH tumors, which are not represented in their study. Furthermore, their calculations do not include shifts in normal-cell heteroplasmies to the tumor cell when assessing selection. Another factor that may explain some of the difference is their use of whole-exome sequencing data, whereas we exclusively use whole-genome sequencing data in order to ensure uniformly deep coverage of the mitochondrial genome. Nonetheless, in the 164 patients for whom WGS data was used for both studies, inferences of heteroplasmy levels of variants are largely concordant (Supplementary Material, Fig. S23). Similarly, Reznik and colleagues (27) examined differences in mtDNA copy number between matched tumor-normal pairs and across tumor samples using TCGA data. Although copy number was not the main focus of our study, we did develop and apply copy number inference methodology that was somewhat different from that used in the Reznik et al. study. MtDNA copy number estimates using the two methods were also quite concordant (Supplementary Material, Fig. S24).
In normal cells, mitochondria are responsible for generating the majority of intracellular energy required for proper cell function. However, tumors preferentially utilize glucose for energy production, a hallmark of cancer (7) (i.e. the Warburg effect (11)). This adaptive tumor environment may remove the cell’s dependence on healthy mitochondria and confer tolerance towards damaged mitochondrial genomes. Damaged mitochondrial genomes can in turn serve as a means to accumulate ROS (1), which play a key role in cancer cell proliferation and survival when present at moderate levels (43). Indeed, we report elevated tumor-specific mutational burden in mitochondrial genes coding for subunits of Complex I, which is the primary source of intracellular ROS production (44). Our study provides evidence that, without reliance on healthy mitochondrial function, tumor cells can utilize mitochondria as vessels for producing tumor-promoting reactive oxygen species.
Mitochondrial involvement in cancer cells has been debated for several decades (11), as the question of whether mitochondria act as drivers or passengers to the tumorigenic process remains unresolved (13,37,45). We suggest a model whereby the tumor generally tolerates neutral drift of most mtDNA alleles, but some alleles confer a selective advantage to the tumor when they rise above a critical threshold (46). Not all disruptive alleles show signals of positive selection, but we posit that this is because excess damage to mtDNA could produce high levels of ROS and induce tumor cell death (43). Along with antioxidant defense to maintain moderate levels of ROS (47), mitochondria may provide an alternative means of control over redox signaling in its earlier stage of production. In order to prevent dangerous amounts of ROS, the tumor cell may hijack the same endogenous mechanisms that are meant to protect mtDNA in normal cells (35,48). Rather than being drivers or passengers to the tumorigenic process, tumor mitochondria may facilitate the process by striking a balance between fully damaged and healthy mitochondrial genomes and consequently maintain tumorigenic levels of ROS. We propose that the retention of a separate mitochondrial genome, which has conferred an evolutionary advantage to eukaryotic cells, may be exploited by tumor cells to promote tumor development and proliferation.
Materials and Methods
Data acquisition and initial quality control
From TCGA’s .bam files, we downloaded (using UCSC CGHub (http://cghub.ucsc.edu; date last accessed August 2015) and TCGA’s BAMSlicer software) reads aligned to the mitochondrial genome and patient-specific two-copy nuclear genes (identified using cbioportal (49)) from whole genome sequencing samples. After processing the samples using SAMtools (50), bam2fastq, and the seqtk software, we then filtered our results to exclude reads that are likely to be nuclear mitochondrial sequences (NuMTs) (Supplementary Material, Fig. S4). NuMTs are DNA sequences that are harbored in the nuclear genome, but closely match sequence in the mitochondrial genome (51). They are likely a result of ancient mitochondrial DNA sequence transfer to the nuclear genome and can lead to false positive variant calls in mtDNA analysis. We filtered NuMTs by removing reads that align with up to one mismatch to the nuclear reference sequence GRCh38 (having removed the mitochondrial revised Cambridge reference sequence, (rCRS) (52)). We then realigned these filtered mitochondrial reads to the GRCh38 human genome build (with rCRS), removed PCR duplicates, and processed each file using GATK’s BaseRecalibrator and INDELRealigner tools (29).
We acquired nuclear somatic mutation data from the UCSC Cancer Genome Browser (https://genome-cancer.ucsc.edu/; date last accessed January 2015) for all relevant TCGA samples. We obtained nuclear mRNA expression values from TCGA Data Portal (https://tcga-data.nci.nih.gov/docs/publicationstcga/; date last accessed August 2015). Where available, we used RNAseq version 2 RPKM values and quantile normalized (53) all RPKM values before downstream analyses. Additionally, we acquired background nuclear somatic mutation rates for TCGA patients from Lawrence et al. (54).
Estimating mtDNA per-cell copy number
To estimate mtDNA copy number across cancer types using WGS data, we calibrated mitochondrial read depth against that of a known two-copy region in the nuclear genome (identified using cbioportal (49)). As described previously (38), we calculated the estimated mtDNA copy number using the formula 2 x (dm/dn), where dm is the average mitochondrial read depth, and dm is the average read depth of the two-copy nuclear gene.
Calling mtDNA variants
Owing to the large numbers of mitochondrial genome copies per cell, an mtDNA variant can exist at levels ranging near-continuously from 0 to 100% (heteroplasmy). In order to accurately call low-level mtDNA variants, we used GATK’s haplotypecaller algorithm, which is able to handle aneuploidy. We found no significant association between number of variants called by GATK and mtDNA copy number (Supplementary Material, Fig. S3). At every mtDNA position, we then classified each sample as either homoplasmic (for either the reference or alternate allele) or heteroplasmic (if the major allele is present at levels below 95%). In normal-cell heteroplasmies, we deemed the allele present in the common ancestor of human and other primates as ‘ancestral’ (22). For each variant, we determined whether it was associated with a haplogroup by applying HaploGrep 2 (55) to the .vcf files. For variants that were only called in the tumor sample under this rubric, we re-inspected the matched normal-cell reads. If any carried the putative mutant allele, we reclassified the variant as a normal-cell heteroplasmy. For cases where GATK reported more than two alleles at a single mtDNA locus, we restricted our analysis to the top two most prevalent alleles in each sample. If a tumor allele was absent in the matched normal, we considered it to be a somatic mutation. We utilized SNPEFF (56) to annotate each variant, while omitting mitochondrial base positions 302-316, 513-526, 566-573, 8860, 16181-16194 from our analysis because they are known to yield erroneous variant calls (23).
Assessing selection on protein-coding genes using dN/dS statistic
We computed ratios of nonsynonymous to synonymous mutations across tumor-specific mutations and normal-cell heteroplasmies. We proceeded to permute these sets of mtDNA variants 1,000 times across coding regions of the mitochondrial genome while maintaining their mutational context (mutated nucleotide residue and its flanking base content) to simulate the null distribution of dN/dS ratios. We then assigned a two-sided P-value and noted the direction of selection signal within normal-cell heteroplasmies and tumor-specific mtDNA mutations across the transcriptome as a whole. Using the same method, we also queried for selective processes on tumor-specific mutations within individual genes and cancer types.
Statistical tests
Unless indicated otherwise, we performed tests for differences in proportions using the R (57) command prop.test, and tests for differences between counts or levels using R’s wilcox.test if the comparison was between two groups and kruskal.test for three or more groups. All reported P-values are for two-sided tests and Bonferroni-corrected if multiple tests are performed, unless stated otherwise. We computed P-values for associations that were adjusted for tumor type and/or age with ANOVA using the anova command in R. We performed gene set enrichment analysis using GSEA (58) version 2-2.2.1 (http://software.broadinstitute.org/gsea/index.jsp; date last accessed January 2016).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. Kaixiong Ye at Cornell University for kindly providing the inferred ancestral human mtDNA sequence. T.L. was supported by American Cancer Society Research Scholar Grant 123436-RSG-12-159-01-DMC, S.G. was supported by NIH NRSA grant F31 CA200349 and NIH training grants T32GM008056-33, Y.N. was supported by R25TCA094186-10. We made use of the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University and the Ohio Supercomputer Center. We also thank Drs. Charis Eng, David Serre, and Angela Ting at the Lerner Genomic Medicine Institute of the Cleveland Clinic for their useful comments on this work.
Conflict of Interest statement. None declared.
Funding
American Cancer Society [123436-RSG-12-159-01-DMC], National Cancer Institute [F31 CA200349] and National Institute of General Medical Sciences [T32 GM008056], National Cancer Institute [R25TCA094186].
References
- 1. Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. (2008) ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science, 320, 661–664. [DOI] [PubMed] [Google Scholar]
- 2. St-Pierre J., Buckingham J.A., Roebuck S.J., Brand M.D. (2002) Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem., 277, 44784–44790. [DOI] [PubMed] [Google Scholar]
- 3. Mattiazzi M., Vijayvergiya C., Gajewski C.D., DeVivo D.C., Lenaz G., Wiedmann M., Manfredi G. (2004) The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum. Mol. Genet., 13, 869–879. [DOI] [PubMed] [Google Scholar]
- 4. Taylor R.W., Turnbull D.M. (2005) Mitochondrial DNA mutations in human disease. Nat. Rev. Genet., 6, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alexeyev M., Shokolenko I., Wilson G., LeDoux S. (2013) The maintenance of mitochondrial DNA integrity–critical analysis and update. Cold Spring Harb. Perspect. Biol., 5, a012641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avital G., Buchshtav M., Zhidkov I., Tuval Feder J., Dadon S., Rubin E., Glass D., Spector T.D., Mishmar D. (2012) Mitochondrial DNA heteroplasmy in diabetes and normal adults: role of acquired and inherited mutational patterns in twins. Hum. Mol. Genet., 21, 4214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanahan D., Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 8. Malik A.N., Czajka A. (2013) Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction?. Mitochondrion, 13, 481–492. [DOI] [PubMed] [Google Scholar]
- 9. Gogvadze V., Zhivotovsky B., Orrenius S. (2010) The Warburg effect and mitochondrial stability in cancer cells. Mol. Aspects Med., 31, 60–74. [DOI] [PubMed] [Google Scholar]
- 10. Kim J.W., Dang C.V. (2006) Cancer's molecular sweet tooth and the Warburg effect. Cancer Res., 66, 8927–8930. [DOI] [PubMed] [Google Scholar]
- 11. Warburg O. (1956) On the origin of cancer cells. Science, 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 12. Koppenol W.H., Bounds P.L., Dang C.V. (2011) Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer, 11, 325–337. [DOI] [PubMed] [Google Scholar]
- 13. Ju Y.S., Alexandrov L.B., Gerstung M., Martincorena I., Nik-Zainal S., Ramakrishna M., Davies H.R., Papaemmanuil E., Gundem G., Shlien A.. et al. (2014) Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pereira L., Soares P., Radivojac P., Li B., Samuels D.C. (2011) Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. Am. J. Hum. Genet., 88, 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li M., Rothwell R., Vermaat M., Wachsmuth M., Schroder R., Laros J.F., van Oven M., de Bakker P.I., Bovenberg J.A., van Duijn C.M.. et al. (2016) Transmission of human mtDNA heteroplasmy in the Genome of the Netherlands families: support for a variable-size bottleneck. Genome Res., 26, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elson J.L., Turnbull D.M., Howell N. (2004) Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet., 74, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freyer C., Cree L.M., Mourier A., Stewart J.B., Koolmeister C., Milenkovic D., Wai T., Floros V.I., Hagstrom E., Chatzidaki E.E.. et al. (2012) Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat. Genet., 44, 1282–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill J.H., Chen Z., Xu H. (2014) Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat. Genet., 46, 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma H., Xu H., O'Farrell P.H. (2014) Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet., 46, 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kennedy S.R., Salk J.J., Schmitt M.W., Loeb L.A. (2013) Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genetics, 9, e1003794.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Greaves L.C., Elson J.L., Nooteboom M., Grady J.P., Taylor G.A., Taylor R.W., Mathers J.C., Kirkwood T.B., Turnbull D.M. (2012) Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genetics, 8, e1003082.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye K., Lu J., Ma F., Keinan A., Gu Z. (2014) Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl Acad. Sci. U.S.A, 111, 10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li M., Schroder R., Ni S., Madea B., Stoneking M. (2015) Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc. Natl Acad. Sci. U.S.A, 112, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samuels D.C., Li C., Li B., Song Z., Torstenson E., Boyd Clay H., Rokas A., Thornton-Wells T.A., Moore J.H., Hughes T.M.. et al. (2013) Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genetics, 9, e1003929.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naue J., Horer S., Sanger T., Strobl C., Hatzer-Grubwieser P., Parson W., Lutz-Bonengel S. (2015) Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion, 20, 82–94. [DOI] [PubMed] [Google Scholar]
- 26. Stewart J.B., Alaei-Mahabadi B., Sabarinathan R., Samuelsson T., Gorodkin J., Gustafsson C.M., Larsson E. (2015) Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genetics, 11, e1005333.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reznik E., Miller M.L., Senbabaoglu Y., Riaz N., Sarungbam J., Tickoo S.K., Al-Ahmadie H.A., Lee W., Seshan V.E., Hakimi A.A.. et al. (2016) Mitochondrial DNA copy number variation across human cancers. eLife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research, N., Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Payne B.A., Wilson I.J., Yu-Wai-Man P., Coxhead J., Deehan D., Horvath R., Taylor R.W., Samuels D.C., Santibanez-Koref M., Chinnery P.F. (2013) Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet., 22, 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levin L., Zhidkov I., Gurman Y., Hawlena H., Mishmar D. (2013) Functional recurrent mutations in the human mitochondrial phylogeny: dual roles in evolution and disease. Genome Biol. Evol., 5, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hurst L.D. (2002) The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet., 18, 486.. [DOI] [PubMed] [Google Scholar]
- 33. Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H., Buhay C., Kang H., Kim S.C., Fahey C.C.. et al. (2014) The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell, 26, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coller H.A., Khrapko K., Bodyak N.D., Nekhaeva E., Herrero-Jimenez P., Thilly W.G. (2001) High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat. Genet., 28, 147–150. [DOI] [PubMed] [Google Scholar]
- 35. Hamers L. (2016) EVOLUTION. Why do cells' power plants hang on to their own genomes?. Science, 351, 903.. [DOI] [PubMed] [Google Scholar]
- 36. Karnkowska A., Vacek V., Zubacova Z., Treitli S.C., Petrzelkova R., Eme L., Novak L., Zarsky V., Barlow L.D., Herman E.K.. et al. (2016) A Eukaryote without a Mitochondrial Organelle. Curr. Biol., 26, 1274–1284. [DOI] [PubMed] [Google Scholar]
- 37. Wallace D.C. (2007) Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Ann. Rev. Biochem., 76, 781–821. [DOI] [PubMed] [Google Scholar]
- 38. McMahon S., Laframboise T. (2014) Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis, 35, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reznik E., Wang Q., La K., Schultz N., Sander C. (2017) Mitochondrial respiratory gene expression is suppressed in many cancers. eLife, 6, e21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harper M.E., Seifert E.L. (2008) Thyroid hormone effects on mitochondrial energetics. Thyroid, 18, 145–156. [DOI] [PubMed] [Google Scholar]
- 41. Volpe A., Novara G., Antonelli A., Bertini R., Billia M., Carmignani G., Cunico S.C., Longo N., Martignoni G., Minervini A.. et al. (2012) Chromophobe renal cell carcinoma (RCC): oncological outcomes and prognostic factors in a large multicentre series. BJU Intl, 110, 76–83. [DOI] [PubMed] [Google Scholar]
- 42. Zong W.X., Rabinowitz J.D., White E. (2016) Mitochondria and Cancer. Mol. Cell, 61, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cairns R.A., Harris I.S., Mak T.W. (2011) Regulation of cancer cell metabolism. Nat. Rev. Cancer, 11, 85–95. [DOI] [PubMed] [Google Scholar]
- 44. Murphy M.P. (2009) How mitochondria produce reactive oxygen species. Biochem. J., 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wallace D.C. (2012) Mitochondria and cancer. Nat. Rev. Cancer, 12, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace D.C., Chalkia D. (2013) Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol., 5, a021220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liou G.Y., Storz P. (2010) Reactive oxygen species in cancer. Free Rad. Res., 44, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Twig G., Hyde B., Shirihai O.S. (2008) Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Et Biophys. Acta, 1777, 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E.. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hazkani-Covo E., Zeller R.M., Martin W. (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet., 6, e1000834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet., 23, 147.. [DOI] [PubMed] [Google Scholar]
- 53. Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- 54. Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A.. et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weissensteiner H., Pacher D., Kloss-Brandstatter A., Forer L., Specht G., Bandelt H.J., Kronenberg F., Salas A., Schonherr S. (2016) HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res., 44, W58–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Venables W.N., Smith D.M., R. and Development Core Team, (2002) An Introduction to R: Notes on R: a Programming Environment for Data Analysis and Graphics, version 1.4.1. Network Theory, Bristol. [Google Scholar]
- 58. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.