Figure 2. Schematic representation of AREs derived from ChIP-Seq and ChIP-exo analyses.
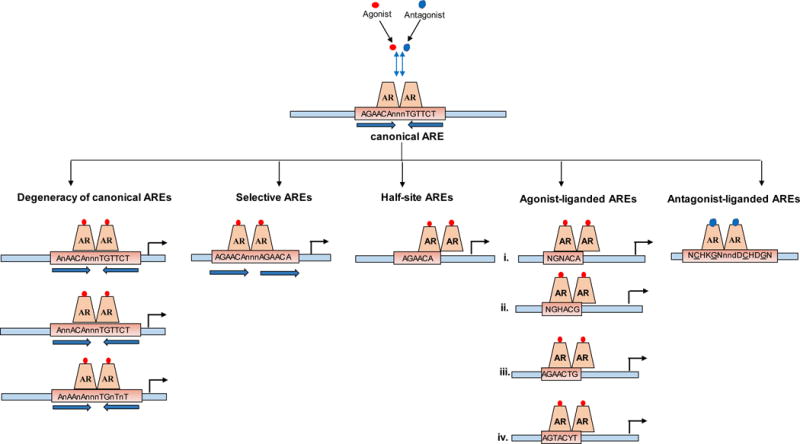
Top panel, consensus sequence for a canonical ARE, which reflects the ideal 15-mer motif of two hexamers arranged in inverted repeat with a 3-mer spacer. Bottom panels, Deviations from this conventional AREs, which include [from left to right]: degenerate AREs in which mismatches of 1 or more basepairs from the consensus canonical ARE motif occur, selective AREs which are composed of direct repeat of 6-mer half-sites, 6-mer half-site ARE sequences found to be enriched in AR binding sites derived from ChIP-based approaches, 4 types of ChIP-exo protected agonist-liganded AREs, ChIP-exo protected antagonist-liganded AREs. Note that, until proven conclusively otherwise, AR is assumed to bind as a dimer to all ARE versions.
