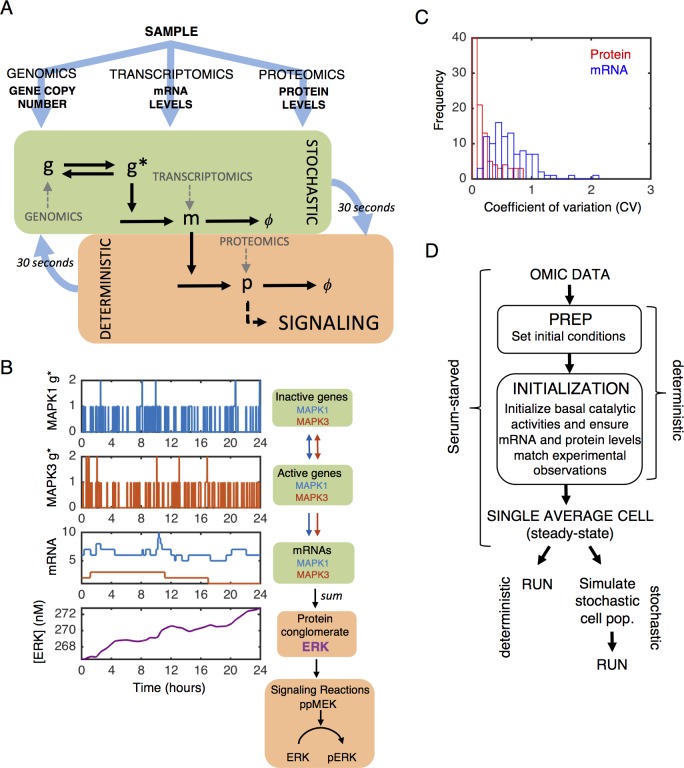
Fig 2. Context tailoring and stochastic gene expression.
A) Gene copy number, mRNA levels, and protein levels inform initial conditions and expression submodel parameters. Gene switching, mRNA synthesis, and mRNA degradation are stochastic, whereas all other processes are deterministic, with a 30 second update interval. B) Example simulations for ERK. Two ERK isoform genes (MAPK1-blue and MAPK3-red; top two plots) switch between active and inactive states, leading to transcription noise (third plot down). Transcripts are summed into the signaling conglomerate ERK (fourth plot down). C) Coefficient of variation (CV) of protein (red) and mRNA (blue) levels across a population of 100 stochastically simulated cells after 24-hour serum-starvation. D) Overview of tailoring and running model. Multi-omic data from serum-starved cells sets initial conditions (“PREP”). Then, a deterministically-simulated initialization procedure maintains consistency with experimental observations in the presence of basal signaling activities resulting in a serum-starved single average cell at steady-state. From there, we run either deterministic or stochastic simulations. Prior to simulating any stimuli for stochastic simulations, we create a population of N number of cells by running the model for a 24-hour period in the serum-starved state an N number of times.