Spinocerebellar ataxias (SCAs) are a heterogeneous group of neurodegenerative disorders that involve the degeneration of the cerebellum and brainstem.1 These genetic diseases are characterized by autosomal dominant inheritance with approximately 44 known subtypes. Recently, dominant mutations in the CACNA1G gene, encoding the voltage-gated calcium channel CaV3.1, have been linked to SCA42 in French2 and Japanese3,4 families. SCA42 prevalence elsewhere in the world has yet to be documented. Through a combination of whole-exome sequencing (WES) and linkage analysis, we have identified an SCA42 mutation in patients from 3 additional countries, expanding the worldwide prevalence of this disease.
Methods
All study methods were approved by the Institutional Review Board of UCLA. All patients received a comprehensive clinical evaluation for acquired causes of ataxia.5 All patients provided written informed consent for collection of DNA. Only patients who tested negative for common genetic ataxias (SCA1, SCA2, SCA3, SCA6, SCA7, and Friedreich ataxia) were enrolled.5 WES was performed for 8 members of 3 families (figure, A and B). Genomic DNA (gDNA) libraries were prepared using the Nextera Rapid Capture Exome kit (Illumina, San Diego, CA). Sequencing for these libraries was performed with 107-bp paired-end reads on a HiSeq 2500 sequencer in the rapid-run mode platform (Illumina, San Diego, CA). The gDNA library for B-III-3 was prepared using the SureSelect Human All Exon V4 Capture kit (Agilent Technologies, Santa Clara, CA), and 101-bp paired-end reads were sequenced on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA). Sequencing data were processed as described.5 Linkage analysis was conducted with ALLEGRO on 190,569 single nucleotide polymorphisms (SNPs) (average distance between SNPs ≈ 0.015 Mb) under a dominant model (f0/f1/f2 = 0/1/1) with estimated minor allele frequencies drawn from HapMap3 project6 Centre d'Etude du Polymorphism Humain - Utah population data, comprised Utah residents with Northern and Western European ancestry.7 Self-reported ancestry was confirmed via principal component analysis (PCA) of WES variants with a minor allele frequency of >5% and 1000 Genomes Project Phase 38 reference data. Segregation analysis for family A was performed by PCR, followed by Sanger sequencing.
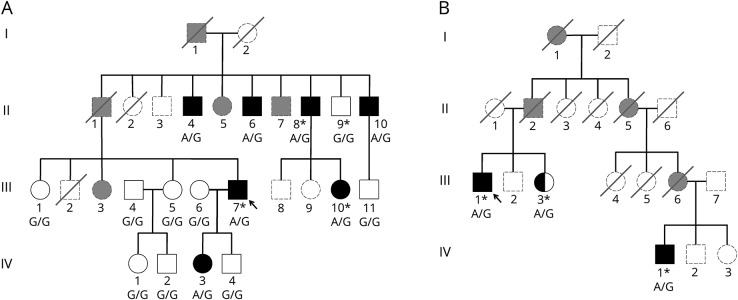
Figure. Pedigrees of the SCA42 families identified in this study.
(A) Pedigree of family A. (B) Pedigree of family B. Probands are indicated by an arrow. Shaded symbols represent affected individuals. Individuals who had WES are indicated by an asterisk. Genotypes of the c.5144 position in CACNA1G are shown under each patient sequenced. G is the reference, and A is the p. R1715H mutation. Individuals who were unavailable for genotyping are represented in gray with dashed lines. Shaded gray individuals were reported as affected. SCA = spinocerebellar ataxia.
Results
Family A was clinically evaluated in Italy, their country of origin, and PCA confirmed European ancestry. Affected members exhibited pure cerebellar ataxia with onset between ages 22 and 58 years (mean 37.5 years). All affected members initially experienced a subjective sense of leg weakness and imbalance that slowly progressed to gait and limb ataxia with dysarthria. There was no nystagmus, abnormal saccadic pursuit, motor deficits, pyramidal or extrapyramidal signs, abnormal sensation, or cognitive decline noted in any patients. MRI of the brain demonstrated cerebellar atrophy in all patients. WES identified a known pathogenic variant in the CACNA1G gene in all 3 affected family members (A-II-8, A-III-10, and A-III-7) tested. All other dominant SCAs were excluded by linkage analysis and/or by the absence of rare missense variation.5 The CACNA1G variant (hg19:chr17:48694921G>A, p.Arg1715His) was previously observed in patients from France and Japan.2–4 This variant was not present in the ExAC (exac.broadinstitute.org) or gnomAD (gnomad.broadinstitute.org) databases of human variation. The variant segregated with disease and was located within a linkage peak, consistent with being pathogenic.
In a second family with pure cerebellar ataxia, family B of Eastern European ancestry living in the United States, the proband (B-III-1) developed, at age 67 years, a sense of imbalance that slowly progressed to a gait and limb ataxia with dysarthria. His first cousin once removed (B-IV-1) was also affected, with an age at onset of 39 years, initially also involving his balance and speech and slowly progressing to a gait and limb ataxia with dysarthria. MRI of the brain showed cerebellar atrophy. PCA verified European ancestry. The above CACNA1G variant was identified in both patients by WES. The proband's sister (B-III-3) also carried the variant but reported herself as asymptomatic and was unavailable for neurologic examination.
Lastly, WES identified the same variant in a man from Yemen, clinically evaluated in Israel, who, at age 25 years, presented with a sense of imbalance and intermittent jerking movements of his neck that slowly progressed to a tremor with cervical dystonia manifesting as a rightward tilt. His imbalance progressed to a mild gait and lower limb ataxia. The remainder of his neurologic examination was normal. MRI of the brain showed mild cerebellar atrophy, primarily of the vermis. His parents and 6 younger siblings were all reported as asymptomatic; however, none were available for examination or genetic testing. Although the 1000 Genomes Project does not include Yemeni samples, this sample clustered with admixed American samples, indicating primarily European ancestry with small amounts of Asian and African admixture, consistent with expectations for a Middle Eastern population.
We next questioned whether CACNA1G variants were common in neurologic disease in the US population, so we reviewed a recent analysis of 3,040 clinical WES cases,9 comprising 1,082 patients with involvement of the CNS, including cerebellar ataxia, and noted that it did not identify any patients with the p.Arg1715His variant we detected and found only 1 patient with a novel variant in CACNA1G deemed pathogenic, suggesting that the mutation of this gene may be rare in the US population. Next, to assess the prevalence of this disease within a specific US ataxia population, we reviewed WES data from 225 sporadic or familial cases seen at our tertiary referral center at UCLA. Aside from family B, we identified no additional cases. Therefore, we estimate the frequency in our ataxia cohort to be 1 of 225 (0.4%) undiagnosed families.
Discussion
In this report, we describe 3 additional SCA42 families originating from Italy, the United States (via Europe), and Yemen, respectively, expanding the worldwide prevalence of the disorder. In addition, we report a case with the new finding of cervical dystonia, along with tremor, previously reported in another family.3 Although previously reported in French and Japanese families,2–4 none of the families in this report had clear evidence of French or Japanese ancestry to the best of our ability to ascertain. Although we find the disorder to be rare in our center's US ataxia population (0.4%, N = 225), we recommend that clinicians broadly consider the possibility of SCA42 in undiagnosed patients with autosomal dominant ataxia, without regard to country of origin.
Author contributions
F.C., G.C., J.E.B., and B.L.F. contributed to the conception and design of the research project and K.J.N., M.A., L.E.P., and J.A.C. were responsible for its execution. A.B.N., M.D.G., S.H.-B., S.P., D.I., A.L., S.C., and B.L.F. supervised collection of the clinical data and samples and/or managed patient care. K.J.N., L.E.P., J.E.B., and B.L.F. conducted all bioinformatics analysis. M.A., K.J.N., and B.L.F. wrote the manuscript, and all authors were responsible for its review and critique.
Study funding
This work was supported in part by the National Institute for Neurological Disorders and Stroke (Grant R01NS082094 to Dr. Fogel) and the National Ataxia Foundation (Young Investigator Award to Dr. Fogel). The research described was supported by the NIH/National Center for Advancing Translational Science UCLA Clinical and Translational Science Institute grant UL1TR000124. Dr. Fogel acknowledges the support through donations to the University of California by the Rochester Ataxia Foundation. MDG was funded by the Michael J. Homer Family Fund. ABN is supported by the Richard and Shirley Cahill Endowed Chair in Parkinson's Disease Research. The authors acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691).
Disclosure
K.J. Ngo, M. Aker, and L.E. Petty report no disclosures. J.A. Chen is a cofounder/advisor of Verge Genomics; has received research support from the NIH/NINDS; and holds stock/stock options in Verge Genomics. F. Cavalcanti reports no disclosures. A.B. Nelson has received research support from the NIH/NINDS, the Parkinson Disease Foundation, the Dystonia Medical Research Foundation, the Brain Research Foundation, and the Richard and Shirley Cahill Foundation. S. Hassin-Baer has received consultancy fees from AbbVie and Medtronic. M.D. Geschwind has received speaker honoraria from Oakstone Publishing, Inc; has served on the editorial board of Dementia & Neuropsychologia; has been a consultant of Advanced Medical Inc, Best Doctors Inc, Grand Rounds, Gerson Lehrman Group Inc, Guidepoint Global, MEDACorp, LCN Consulting, Optio Biopharma Solutions, various medical-legal consulting, Biohaven Pharmaceuticals Inc, Teva Pharmaceuticals, and Quest Diagnostics; and has received research support from the NIH, the Michael J. Homer Family Fund, CurePSP, and the Tau Consortium. S. Perlman has received research support from EryDel, Reata, Viropharma/Shire, Retrotope, Edison, Teva, Pfizer, Biohaven, and the National Ataxia Foundation. D. Italiano and A. Lagana report no disclosures. S. Cavallaro has served on the editorial boards of the ARC Journal of Neuroscience, Current Medicinal Chemistry, Genes and Genome Journal, ISRN Genomics, Jacobs Journal of Biomarkers, Journal of Biological Informatics & Biodiversity, Journal of Genomes and Proteomes, Letters in Drug Design & Discovery, Medicinal Chemistry, The Open Biochemistry Journal, and Source Journal of Genomics. G. Coppola receives publishing royalties from Oxford University Press and has received research support from Takeda Pharmaceutical Company, the NIH, the Adelson Medical Research Foundation, the Tau Consortium, the CHDI, Takeda Pharmaceutical Company, the John Douglas French Alzheimer's Foundation, and the FARA. J.E. Below has received research support from the Sanofi Innovation Awards Program, the Tulane National Primate Research Center, the NIH/Wayne State University Genetic Study of Stuttering, the NIH/Johns Hopkins University Baylor-Johns Hopkins Center for Mendelian Genetics, and the NIH/Broad Institute of MIT. B.L. Fogel has received travel funding/speaker honoraria from the American Academy of Neurology, the American Physician Institute for Advanced Professional Studies, and the National Ataxia Foundation; has served on the editorial boards of Neurology: Genetics and Neurology Today; and has received research support from the NIH/NINDS and the National Ataxia Foundation. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Shakkottai VG, Fogel BL. Clinical neurogenetics: autosomal dominant spinocerebellar ataxia. Neurol Clin 2013;31:987–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coutelier M, Blesneac I, Monteil A, et al. . A recurrent mutation in CACNA1G alters cav3.1 T-type calcium-channel conduction and causes autosomal-dominant cerebellar ataxia. Am J Hum Genet 2015;97:726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morino H, Matsuda Y, Muguruma K, et al. . A mutation in the low voltage-gated calcium channel CACNA1G alters the physiological properties of the channel, causing spinocerebellar ataxia. Mol Brain 2015;8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura M, Yabe I, Hama Y, et al. . SCA42 mutation analysis in a case series of Japanese patients with spinocerebellar ataxia. J Hum Genet 2017;62:857–859. [DOI] [PubMed] [Google Scholar]
- 5.Fogel BL, Lee H, Deignan JL, et al. . Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol 2014;71:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International HapMap C; Altshuler DM, Gibbs RA, et al. . Integrating common and rare genetic variation in diverse human populations. Nature 2010;467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000;25:12–13. [DOI] [PubMed] [Google Scholar]
- 8.Genomes Project C, Auton A, Brooks LD, et al. . A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Retterer K, Juusola J, Cho MT, et al. . Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016;18:696–704. [DOI] [PubMed] [Google Scholar]