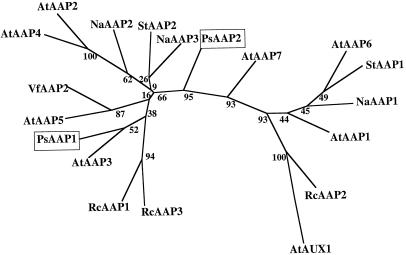
Figure 3.
Computer-aided analysis of homologies between PsAAP1 and PsAAP2 and related proteins. The analysis was performed using PHYLIP (J. Felsenstein, 1995, Phylogenetic Interference Package, version 3.5, distributed by the author: Department of Genetics, University of Washington, Seattle, WA) with aligned sequences of PsAAP1 and PsAAP2 and amino acid transporters from Arabidopsis (AtAAPs), S. tuberosum (StAAPs), R. communis (RcAAPs; RcAAP3-accession no. AJ132228) (Fischer et al., 1998), N. alata (NaAAPs; Schulze et al., 1999), and V. faba (VfAAP2, accession no. Y09591). The numbers indicate the occurrence of a given branch in 100 bootstrap replicates of the given data set. AtAUX1 was used as the outgroup.