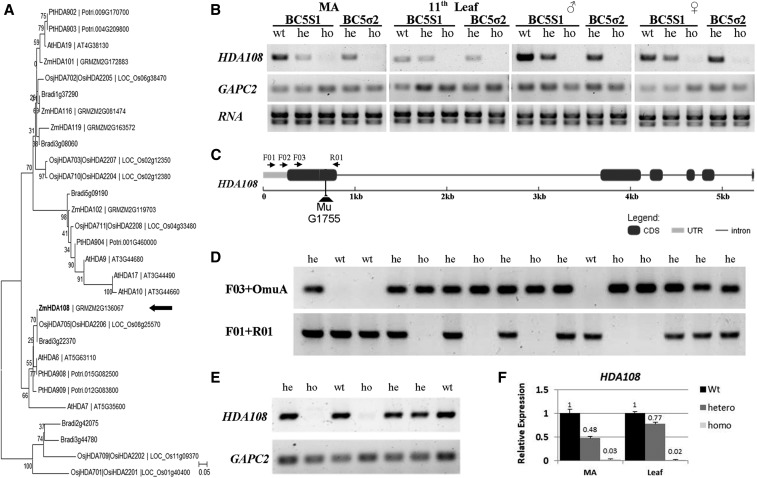
Figure 1.
HDA108 gene structure, diagram of MuG175 insertion, plant genotyping, and expression analysis. (A) Neighbor-Joining phylogenetic tree of RPD3/HDA1 histone deacetylases of Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa (spp. japonica and indica-cultivar group), Populus trichocarpa, and Zea mays (accessions and sequences of the HDA proteins are reported in File S10). The tree is based on a Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) multiple alignment and was reconstructed using MEGA7; bootstrap was inferred from 1000 replicates. (B) RT-PCR expression analysis of HDA108 in the two segregating progenies BC5S1 and BC5σ2 with different genotypes: wild-type (wt) heterozygous (he) homozygous (ho) in meristematic enriched tissues (MA), basal part of 11th leaf, male (♂) and female (♀) inflorescence. HDA108 is expressed in the wild-type tissues but is not expressed in homozygous genotypes. A lower level of expression characterized the heterozygous genotypes. (C) HDA108 gene structure showing exon/intron structure, Mu G1755 insertion, and primer position and orientation. (D) Genotyping of a BC5S1 progeny showing the different PCR amplification patterns of the three possible genotypes (wt: wild-type; he: heterozygous; and ho: homozygous) identified with two primer combinations: F03-OmuA (OmuA is designed on the Mu inverted repeats) and F01-R01. (E) RT-PCR expression analysis on the sixth leaf showing that HDA108 transcript is not detected in homozygous mutant plants. (F) Quantitative Real-Time PCR expression analysis in meristematic enriched tissues (MA) and leaves confirms the lack of HDA108 transcript in homozygous tissues (34- and 55-fold decrease in transcript level, respectively, as compared to wild type) and the intermediate expression level in the heterozygous samples.