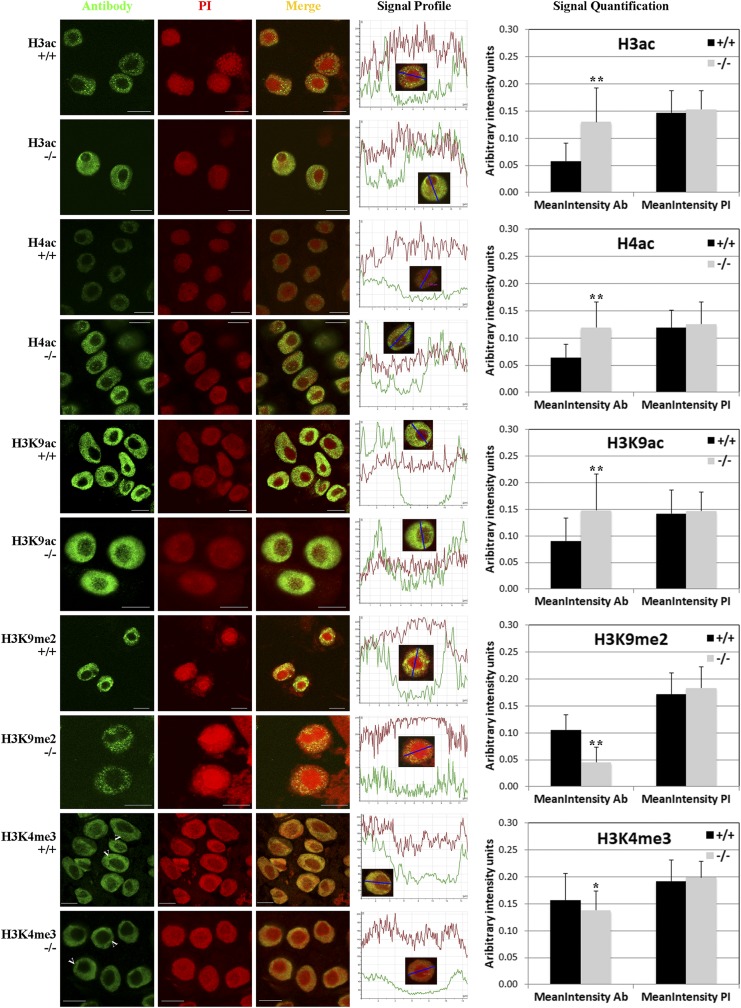
Figure 3.
Effect of had108 knockout on total levels and nuclear distribution of histone H3 and H4 modifications. Nuclei from wild-type (+/+) and homozygous (−/−) hda108 mutant plants were immunostained with anti-H3ac, H4ac, H3K9ac, H3K9me2, and H3K4me3 antibodies to analyze the nuclear distribution and levels of modified histones (green signal) and counterstained with propidium iodide (PI; red signal). A minimum of 50 nuclei from wild-type and homozygous mutant root apexes in three replicated immunolocalizations were observed. For each antibody × genotype combination, the signal profiles of fluorescence emission throughout the nucleus, obtained by Leica Confocal Software, are reported for both antibody and propidium iodide. Profile y-axis shows the fluorescence signal intensity (arbitrary units) measured along the line segment (reported on x-axis; µm) shown in the inset nucleus. On the right, signal quantification graphs report the mean nucleus intensities (arbitrary units) calculated for both antibody and propidium iodide signal in each genotype, using the CellProfiler automated image analysis software. For each antibody × genotype combination, at least 200 nuclei (on different slides of the three replicates) were selected and the mean nucleus intensities calculated for both antibody and propidium iodide signal. Summary data from image analyses are reported in File S1. Bars represent SD. Asterisks indicate statistically significant changes: * P ≤ 0.05, ** P ≤ 0.01; Student’s t-test. Arrowheads point to the chromocenters. Bar, 10 µm.