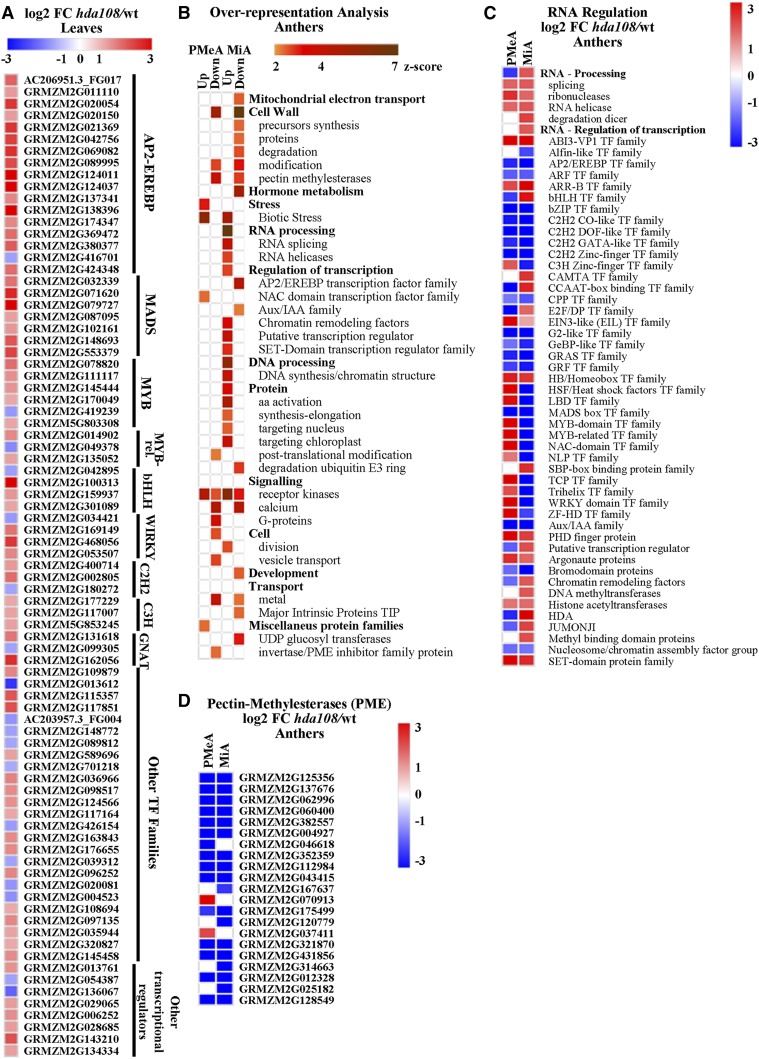
Figure 5.
Pathway and gene family analysis of hda108 differentially expressed genes. (A) Log2 fold change heat map of transcription factor coding genes differentially expressed in mutant leaf. TFs were identified according to MapMan (Thimm et al. 2004; Usadel et al. 2009) and Grassius database (http://grassius.org/) annotations. (B) MapMan functional categories enriched in hda108 up- and down-regulated genes in postmeiotic (PMeA) and mitotic (MiA) anthers. Z-scores automatically calculated from P-values (e.g., 1.96 corresponds to a P-value of 0.05) are plotted in an orange to brown color scale. (C) Log2 fold change heat map of gene families included in the “RNA processing” and “RNA regulation of transcription” categories that are differentially expressed in hda108 PMeA or MiA. The Morpheus (https://software.broadinstitute.org/morpheus/) “Collapse” tool was used to aggregate DE genes belonging to the same gene family and to produce the heat map. (D) Log2 fold change heat map of gene annotated as pectin-methylesterases (PME) and differentially expressed in hda108 PMeA or MiA.