Fig. 2.
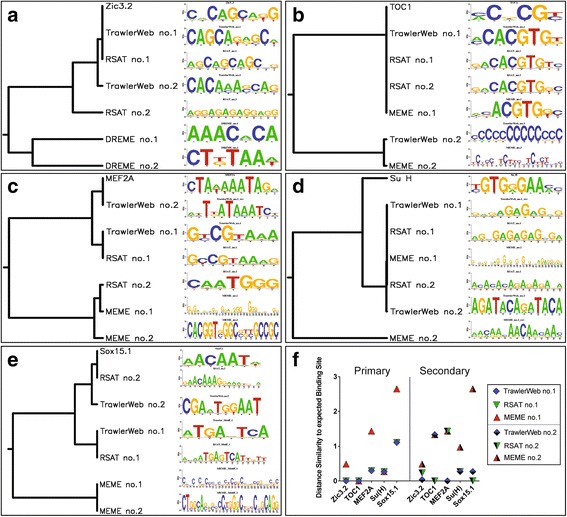
Finding the expected motif with TrawlerWeb, RSAT peak-motifs, and MEME-ChIP. Alignment of the closest primary (no.1) and secondary (no.2) motif to the expected binding site identified for each motif discovery tool for the five species a Danio rerio, b Arabidopsis thaliana, c Mus musculus, d Drosophila melanogaster, and e Homo sapiens. f For each tool, Similarity Distance of the closest primary (no.1) and secondary (no.2) motif to the expected binding site. Motifs of 6 nucleotides (nt) length were represented for Su(H) and Sox15.1, and 7 nt for MEF2A, TOC1, and Zic3.2. MEME did not find any motif for Zic3.2, motif found by DREME was used
