Fig. 4.
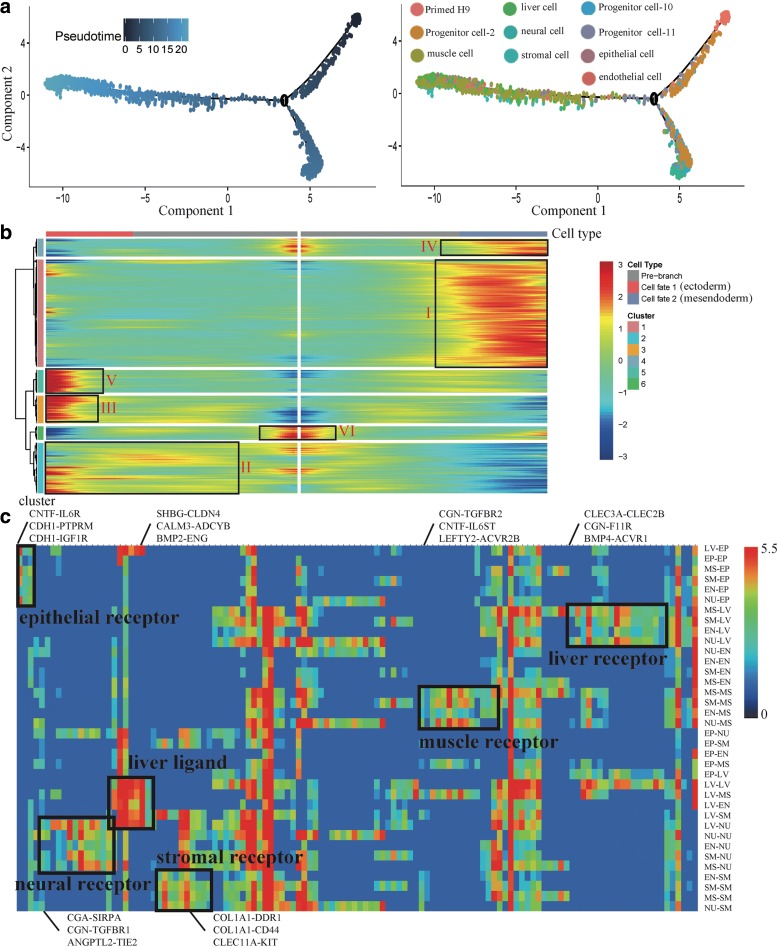
EBs simulate the early development in vivo. a Differentiation trajectory of EBs constructed by Monocle. b Heatmap shows the gene expression dynamics during EB differentiation. Genes (row) are clustered and cells (column) are ordered according to the pseudotime development. Genes are listed in Additional file 6: Table S5. Gene clusters I–VI were selected for further analysis. c Heatmap shows the mean number of cell–cell interactions. LV liver cell, EP epithelial cell, MS muscle cell, SM stromal cell, EN endothelial cell, NU neural cell. List of ligand-receptor pairings (column) and cell–cell pairings (row) are listed in Additional file 7: Table S6