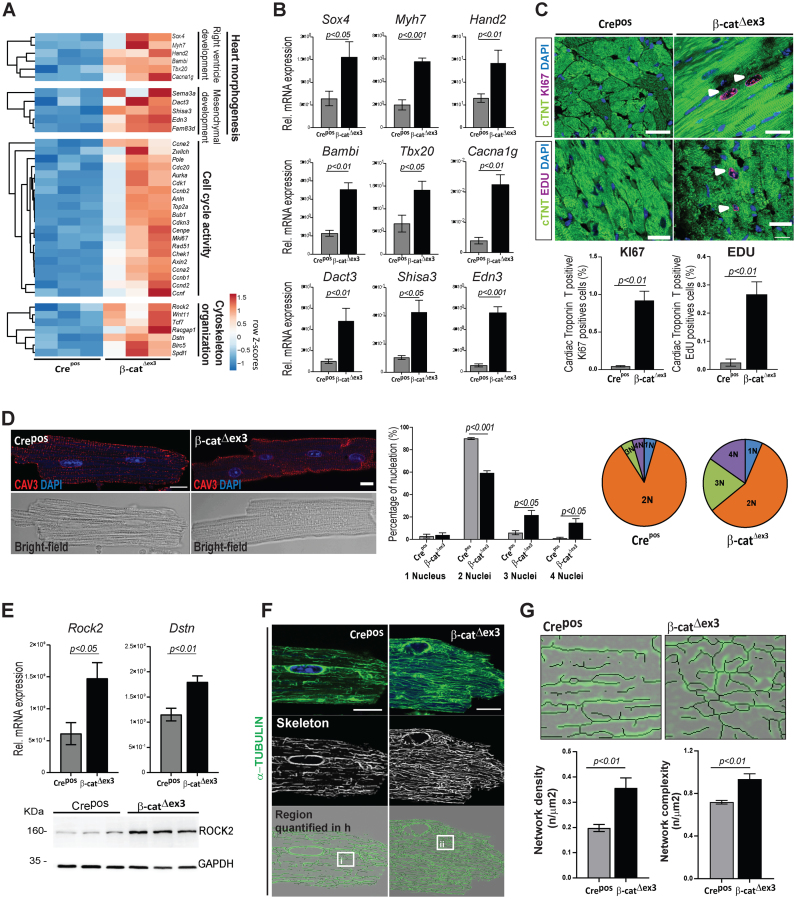
Figure 3.
Wnt transcriptional activation results in increased CM cell cycling and cytoskeletal remodelling in the adult heart. (A) Heatmaps depicting row Z-scores of RPKMs of upregulated genes involved in heart morphogenesis, cell cycling and cytoskeleton organization in β-catΔex3 ventricles; with (B) corresponding qPCR validations (β-catΔex3n = 4; Crepos n = 5). (C) Confocal images of KI67 or EdU (magenta) immunostainings with quantification of double positive cTNT/KI67 and EdU cells (n = 3; >400 cells/mouse). White arrows indicate KI67 and EDU positive CM. (D) Immunostainings for Caveolin 3 (red), DAPI (blue) in single CM along with quantification of nuclei/CM (n = 3; ≥90 cells/CM per mouse). (E) qPCR validating increased cytoskeletal regulators Rock2 and Dstn; and immunoblot showing increased ROCK2 in β-catΔex3 ventricles (β-catΔex3n = 4; Creposn = 5). (F) Confocal images and (G) quantification of microtubule network density and complexity in single CMs (n = 3; 5–8 cells/mouse; i-ii represent higher magnification in F). Images are an overlay of α-TUBULIN images (green) with corresponding extracted skeletons (black). Tbp was used for normalization in B and E and GAPDH was loading control in F. Scale bar C: 20 μm, E, G: 10 μm. Confocal images were re-colored for color-safe combinations.