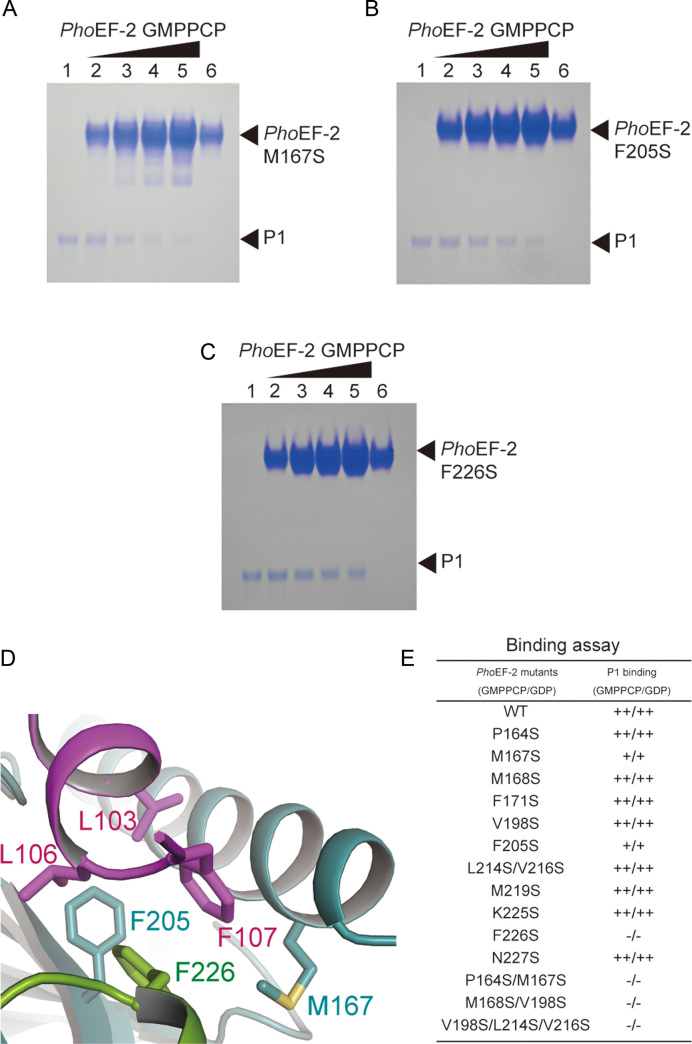
Figure 4.
P1-binding analysis of PhoEF-2 using point mutants. (A–C) are results for GMPPCP-bound PhoEF-2 mutants M167S, F205S, and F226S, respectively. The homodimer of P1 (100 pmol) was incubated without PhoEF-2 mutants (lane 1), or with 100 pmol (lane 2), 200 pmol (lane 3), 300 pmol (lane 4) or 400 pmol (lane 5) of the PhoEF-2 mutants in 10 μl of solution at 70°C. Each PhoEF-2 mutant (100 pmol) was also incubated without P1 (lane 6). (D) Detailed view of the interaction between M167, F205, and F226 of PhoEF-2 and L103, L106, and F107 of P1C11. The side chains of these residues are represented by stick models. (E) Comparison of the P1-binding ability of GMPPCP/GDP-bound PhoEF-2 mutants. The binding ability of each mutant is displayed as ++ (comparable to WT), + (less closely than WT) or – (undetectable).