Figure 5.
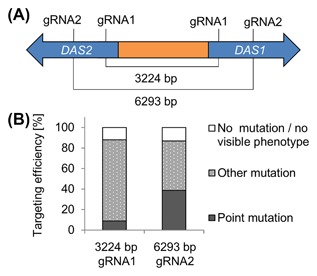
Dual cutting gRNAs for DNA removal. A, The P. pastoris DAS1/DAS2 locus consisting of DAS1 and DAS2, which are located in the reverse complement orientation and are interspaced with the respective promoters and HOB3, encoding a putative guanosine nucleotide exchange factor. Due to 91% sequence identity a single gRNA can be designed to introduce double strand breaks in the DAS1 and DAS2 gene. B, P. pastoris CBS 7435 wildtype was transformed with CRISPR‐Cas9 plasmids bearing HsCas9 and either DAS1/DAS2‐gRNA1 or DAS1/DAS2‐gRNA2. Transformants were cultivated in 96‐DWPs and then stamped on minimal media plates containing glucose (BMD1) and methanol (BMM1) as carbon source to determine the targeting efficiencies. The type of mutation was elucidated by sequencing of >10 methanol deficient transformants/construct. Mean values and standard deviations of biological triplicates are shown. Increasing the distance of the targeting sites increases the frequencies of NHEJ mutations and reduces the probability for sequence removal
