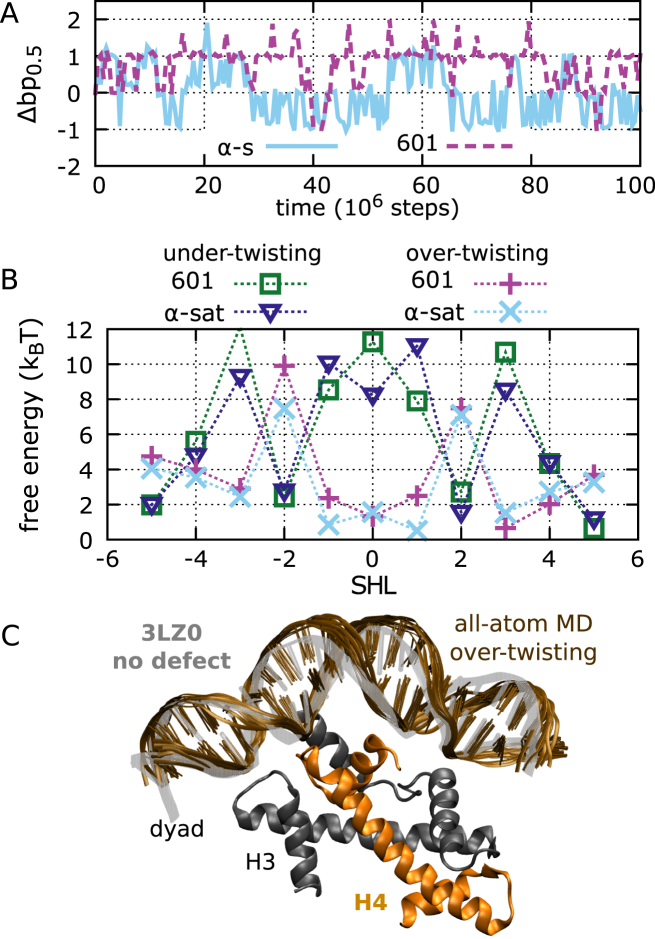
Figure 7.
Stability and defect formation in positioning sequences. (A) The dynamics of contact index 0.5 for 601 and α-satellite nucleosomes for two representative trajectories, starting from the optimal conformations found in the respective crystal structures. (B) The free-energy costs of defect formation at each SHL for 601 and α-satellite positioning sequences. (C) Structure of 601 nucleosomes around SHL –1, comparing the crystal structure with PDB id 3LZ0 (gray transparency) to the metastable form with over-twisting (brown). For the latter, we show 10 DNA frames sampled from our 100-ns all-atom MD simulation starting from a configuration back-mapped from a coarse-grained trajectory. We also show the region of the H3H4 tetramer involved in the interactions with the DNA, highlighting that the extra base pair is accommodated in the outer DNA region lacking histone contacts.