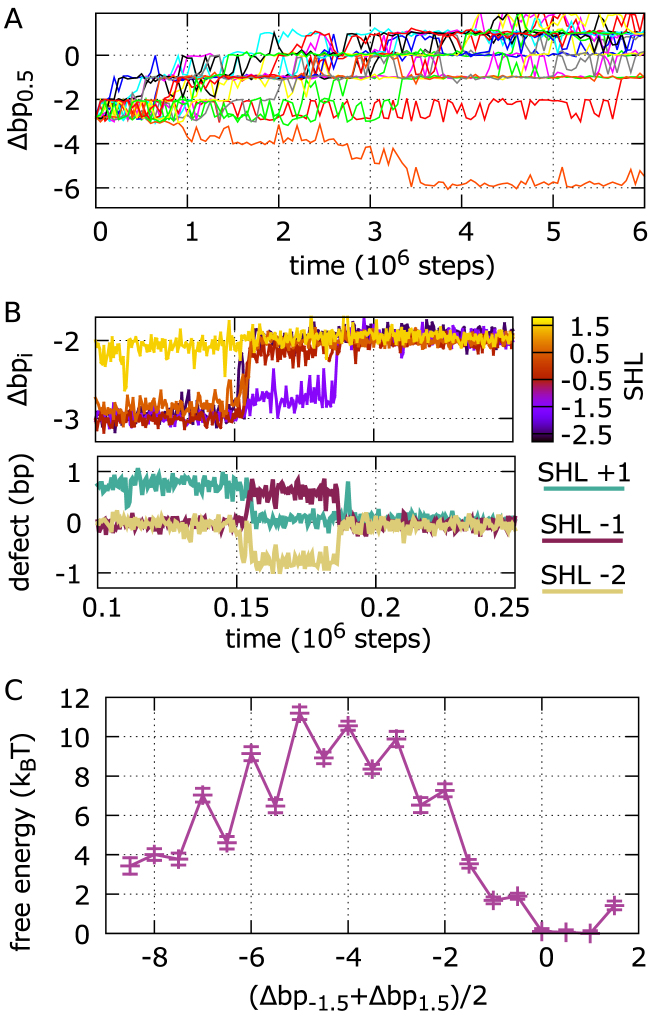
Figure 8.
Repositioning of 601 nucleosomes via defect formation. (A) Dynamics of contact index 0.5 for a sample of 20 trajectories starting from an initial configuration shifted by 3 bp from the crystal structure. In most trajectories, repositioning proceeds toward the optimal crystal structure configuration with contact indexes zero; with this shifted initial condition, out of a total of 400 MD runs, we only find 3 sliding events going in the opposite direction. (B) Dynamics of the central contact indexes during a section of a sliding trajectory (top), showing that repositioning occurs via formation or disappearance of defects at SHLs –2, –1 and +1 (bottom). (C) Free energy profile of 601 repositioning reconstructed via Markov state modeling, as a function of the average of the contact indexes at SHLs ±1.5. Integer states have no defects, while half-integer ones have DNA over-twisting at SHLs 0, –1 or +1.