Abstract
The muscleblind-like (MBNL) family of proteins are key developmental regulators of alternative splicing. Sequestration of MBNL proteins by expanded CUG/CCUG repeat RNA transcripts is a major pathogenic mechanism in the neuromuscular disorder myotonic dystrophy (DM). MBNL1 contains four zinc finger (ZF) motifs that form two tandem RNA binding domains (ZF1–2 and ZF3–4) which each bind YGCY RNA motifs. In an effort to determine the differences in function between these domains, we designed and characterized synthetic MBNL proteins with duplicate ZF1–2 or ZF3–4 domains, referred to as MBNL-AA and MBNL-BB, respectively. Analysis of splicing regulation revealed that MBNL-AA had up to 5-fold increased splicing activity while MBNL-BB had 4-fold decreased activity compared to a MBNL protein with the canonical arrangement of zinc finger domains. RNA binding analysis revealed that the variations in splicing activity are due to differences in RNA binding specificities between the two ZF domains rather than binding affinity. Our findings indicate that ZF1–2 drives splicing regulation via recognition of YGCY RNA motifs while ZF3–4 acts as a general RNA binding domain. Our studies suggest that synthetic MBNL proteins with improved or altered splicing activity have the potential to be used as both tools for investigating splicing regulation and protein therapeutics for DM and other microsatellite diseases.
INTRODUCTION
Alternative splicing (AS) is a complex and versatile process of post-transcriptional gene regulation whereby exons within a precursor RNA transcript are differentially joined and introns removed to produce a mature mRNA. AS generates multiple mRNA isoforms from an individual gene most often resulting in the expression of a diverse set of protein products. Additionally, AS alters the fate of mRNAs through the inclusion of regions that impact RNA localization, translation, and turnover (1). It is now recognized due to large-scale transcriptome based studies that more than 90% of human protein coding genes undergo AS, making regulation of this process critical for proper cellular function (2,3). Trans-acting protein factors, including RNA binding proteins (RBPs, reviewed in (4) and (5)), can function as regulators of AS by interacting with specific RNA motifs, or splicing regulatory elements, to enhance or repress the inclusion of alternative exons. RBPs also act in a spatio-temporal and developmentally dependent manner to modulate the overall profile of mRNAs produced within specific cell types, developmental stages, or in response to varying environmental conditions (1,6).
Muscleblind-like (MBNL) proteins are a family of highly conserved RBPs that regulate RNA metabolism during tissue-specific development, most notably the activation or repression of alternative exon inclusion (7,8). MBNL proteins have been specifically implicated in regulating fetal to adult mRNA isoform transitions in heart and muscle (9–13). In addition, MBNL proteins have been linked to the regulation of other RNA metabolic processes including localization (14), turnover (15), gene expression (16,17), alternative polyadenylation (18) and micro-RNA processing (19).
MBNL proteins, particularly MBNL1, have been the focus of intense study for the past 15 years due to their prominent role in the pathogenesis of myotonic dystrophy (DM). DM is a multi-systemic neuromuscular disorder caused by expression of CTG or CCTG repeat expansions within the 3′ untranslated region of DMPK (DM Type 1) or intron 1 of CNBP (DM Type 2), respectively (20,21). Once transcribed into RNA, these expanded CUG or CCUG repeats sequester MBNL proteins into discrete nuclear RNA-protein aggregates called foci (22–24). Sequestration of MBNL by these toxic, expanded RNAs leads to dysregulation of MBNL-mediated AS linked to causing some of the disease symptoms (25–28). Although most commonly associated with DM, loss of MBNL1 function has also been associated with other disorders, specifically spinocerebellar ataxia type 8 (SCA8) and Fuchs Endothelial Corneal Dystrophy (FECD) (29,30).
In order to regulate specific splicing events, MBNL1 acts as an enhancer or repressor of exon inclusion in a transcript dependent manner. In general, if MBNL1 binds upstream of a regulated exon it suppresses inclusion and if it binds downstream it enhances inclusion (17,31,32). RNA binding by MBNL1 is mediated via four highly conserved CCCH-type (CX7CX4–6CX3H) zinc finger (ZF) motifs that fold into two tandem RNA binding domains commonly referred to as ZF1–2 and ZF3–4 (31,33). These two domains are located within the N-terminal region of the protein and are separated by a flexible linker predicted to mediate MBNL1 binding to a wide variety of RNAs (33,34). Studies have shown that MBNL proteins bind YGCY (Y = C or U) motifs within their RNA targets (32,35). Crosslinking immunoprecipitation (CLIP)-seq and RNA Bind-n-seq (RBNS) experiments have identified several additional related motifs (14,36). The expanded CUG / CCUG repeat RNA in DM patients contain many YGCY motifs, providing a sink for MBNL and the subsequent dysregulation of RNA processing mediated by MBNL proteins.
Sequence alignment and secondary structural overlay of the two ZF domains show that ZF1 / ZF3 and ZF2 / ZF4 have high sequence similarity and nearly identical structures (31,33,37). The major differences between the domains is (i) an extended α-helix at the end of ZF2, (ii) an interdomain linker that is two amino acids shorter in the ZF1–2 domain (Supplemental Figure S1), and (iii) a short N-terminal helix before ZF1 absent in the ZF3–4 domain (31,33,37). Due to the high degree of similarity between the two domains as well as their physical separation via the linker, it has been predicted that the ZF domains have the same or very similar RNA binding activities and may be functionally redundant (31,33). The hypothesis of functional redundancy is further supported by studies with the Drosophila melanogaster and Caenorhabditis elegans orthologs of the MBNL1 gene, muscleblind (mbl). Mbl from these organisms contains only a single ZF domain or the major isoform contains only a single ZF domain orthologous to the human ZF1–2 and yet is able to regulate splicing of many MBNL1 target transcripts in mammalian cell culture (38–40).
Despite the similarity between these domains, combinatorial mutagenic analysis of the four ZFs found that ZF1–2 and ZF3–4 are not functionally equivalent (31). Using this approach it was discovered that a MBNL1 protein with a single functional ZF1–2 bound with higher affinity to all tested RNA substrates compared to a MBNL1 with only a functional ZF3–4 (31). Additionally, a MBNL1 with only an active ZF1–2 retained approximately 80% of splicing activity while the MBNL1 mutant with only an active ZF3–4 maintained 50% splicing regulation (31). Despite these observations it still remained unclear if the ZF pairs truly act as independent domains. Additionally, the function of the individual ZF domains and whether they cooperate in some manner through higher-order interactions to achieve AS regulation remained ambiguous.
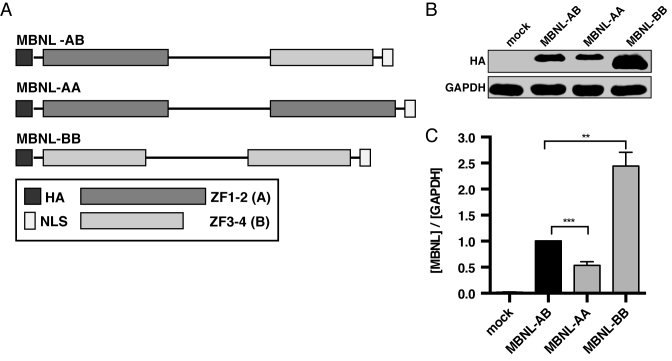
In order to address these questions, we utilized a synthetic biology approach to generate chimeric MBNL1 proteins with novel ZF domain organization. Specifically, we hypothesized that a synthetic MBNL1 protein with higher RNA binding affinity and subsequent splicing activity could be engineered by replacing ZF3–4 with a second ZF1–2. Additionally, we predicted that substitution of the ZF1–2 domain with a ZF3–4 would result in weakened RNA binding and reduced splicing regulation. To test these hypotheses, two synthetic MBNL constructs were designed with duplicate ZF domains: (i) a MBNL in which the ZF3–4 domain is replaced with a ZF1–2 (defined as domain A) to create MBNL-AA and (ii) MBNL-BB in which the ZF1–2 domain is substituted with a ZF3–4 (defined as domain B) (Figure 1A).
Figure 1.
Zinc finger domain architecture and protein expression levels of synthetic MBNL proteins. (A) Schematic of synthetic MBNL proteins that shows organization of zinc finger domains (ZF1–2 (Domain A) and ZF3–4 (Domain B)) and location of HA tag and nuclear localization signal (NLS). The length of the individual segments are proportional to the size of each region of the protein. (B) Representative immunoblot comparing relative protein levels of synthetic MBNL proteins in transfected HeLa cells. (C) Quantification of synthetic protein levels in HeLa cells via western blot against the HA tag (n = 4). Relative levels of each protein were normalized to GAPDH. MBNL-AB expression values were then set equal to 1 and MBNL-AA and MBNL-BB protein levels normalized (data represented as mean ± standard error; **P < 0.01, ***P < 0.001, Student's t-test).
Using this approach we discovered that the ZF1–2 and ZF3–4 domains act as independent units with distinct characteristics, most notably different RNA binding specificities. We also showed that the ZF domains can be organized in novel ways to produce synthetic MBNL1 proteins with different activities as assayed by AS and RNA binding assays. The creation and characterization of these synthetic proteins has not only given us additional insights into the function of the individual ZF domains, but also provides a framework to develop novel MBNL proteins with the potential to serve as tools to investigate AS regulation and act as therapeutic biologics for DM and other microsatellite diseases.
MATERIALS AND METHODS
Protein design, synthesis and cloning
The wild-type (WT) MBNL1 protein (amino acids 1–382; splice isoform a; NCBI accession number NP_066368) was used as a template for the construction of the MBNL-AB, MBNL-AA and MBNL-BB constructs. Due to the difficulty of purifying MBNL1 with the C-terminal region (amino acids 261–382) and to reduce the size of our synthetic proteins, we chose to exclude this portion of the protein in our synthetic design. Previous studies have shown that the C-terminal region is not required for high-affinity RNA binding (41,42). MBNL-AB was created using primers to add the N-terminal HA tag and the C-terminal nuclear localization signal (SV40 NLS). The sequence of MBNL-AA and MBNL-BB was synthesized (GenScript). All three proteins were cloned into pCI (Promega) for mammalian expression and pGEX-6P-1 (Amersham) for bacterial protein expression using XhoI and NotI sites. The amino acid sequences of all MBNL constructs are reported in Supplemental Figure S1A.
Creation of stable, inducible synthetic MBNL expression cell lines
N-terminal GFP-tagged constructs encoding MBNL-AB and MBNL-AA with the HA tag removed were cloned into PB-PuroTet, a vector containing PiggyBac Transposon sequences (43) flanking a PGK-driven puromycin cassette and a minimal CMV promotor downstream of a TetR response element (TRE) to drive doxycycline-inducible expression of the GFP-MBNL construct. The In-Fusion cloning system (Clonetech) was utilized according to the manufacturer's instructions to clone the GFP-tagged constructs into the PB-PuroTet vector. At 60% confluency in six-well plates, mbnl 1/2 double knockout mouse embryonic fibroblasts (MEFs), gifted by Maurice Swanson, were transfected with 1 μg of PB-PuroTet vector encoding GFP tagged MBNL-AB or MBNL-AA, 1 μg of a PB-Tet-On Advanced (vector containing PiggyBac Transposon sequences (43) flanking rtTA Advanced (Clontech) under CMV-driven expression as well as a puromycin selection cassette), and 1 μg of PiggyBac transposase (total = 3 μg) using TransIT-LT1 (Mirus) as per the manufacturer's instructions. After 24 h, the cells were subjected to puromycin selection (4 μg/ml), allowed to recover for several days, and then exposed to 1000 ng/ml doxycycline (Sigma) for 24 h. Cells were then sorted for high GFP expression using the SH800S Cell Sorter (Sony). Individual clones were isolated and the populations expanded in the presence of puromycin. Individual clones for each cell line were selected for experimental use based on GFP-MBNL expression across a range of doxycycline concentrations.
Cell culture and transfection
HeLa cells were cultured as a monolayer in Dulbecco's modified Eagle's medium (DMEM)-Glutamax (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1X antibiotic-antimycotic (Gibco) at 37°C under 5% CO2. Prior to transfection, cells were plated in twelve-well plates at a density of 8 × 104 cells/well. Cells were transfected approximately 36 h later at ∼80% confluency. Plasmids (400 ng/well) were transfected using 2 μl of Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. Cells were placed in Opti-MEM I reduced serum media (Gibco) at the time of transfection. Six hours later, the Opti-MEM I was replaced with our supplemented DMEM. 18 hours post-medium exchange cells were harvested using TrypLE (Gibco) and pelleted using centrifugation.
For overexpression cell-based splicing assays (Figure 2) and Western blots (Figure 1B), 200 ng of protein plasmid or empty pCI vector (mock) were co-transfected with 200 ng of minigene. In the context of the plasmid dosing system (both splicing assays and Western blots) (Figure 3 and Supplemental Figure S6), 200 ng of a selected minigene was co-transfected with increasing amounts of protein expression plasmid up to 200 ng. In cases where less than 200 ng were transfected, empty pCI vector was used to make up the remainder of the total 400 ng transfected. When plasmid dosing was performed in the context of CUG repeat RNA (Figure 7), the amount of protein expression vector remained unchanged from previous dosing experiments, but only 100 ng of the selected minigene was transfected with 100 ng of a DMPK-CUG960 expressing plasmid (34).
Figure 2.
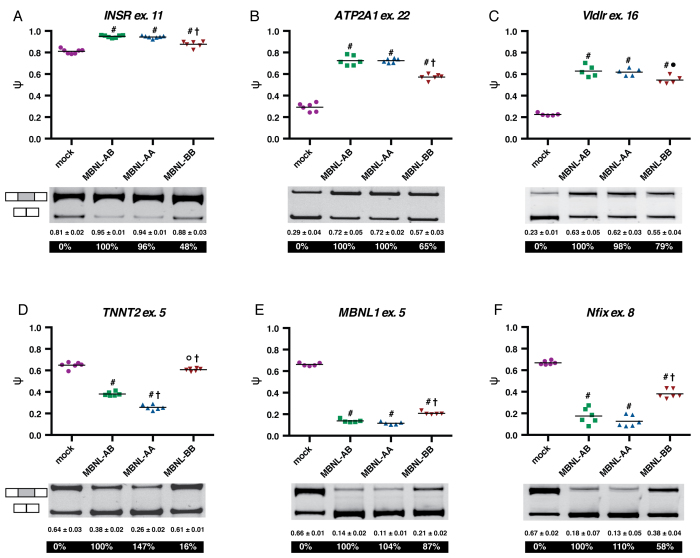
Synthetic MBNL proteins regulate splicing of minigenes in HeLa cells with different activities. (A–F) Jitter plot representations of cell-based splicing assays using INSR, ATP2A1, Vldlr, TNNT2, MBNL1, and Nfix minigenes, respectively. HeLa cells were transfected with empty vector (mock) or MBNL protein expression plasmids and a single minigene reporter. Percent spliced in (PSI, ψ) (i.e. percent exon inclusion) for each protein treatment was then quantified. Each point is from a single experiment and the line represents the average of all experiments for that condition (at least n = 5 for each protein treatment). Average ψ (± standard deviation) and percent splicing activity (displayed in white) are listed below the representative splicing gels. (oP < 0.05 versus mock, #P < 0.0001 versus mock, •P < 0.05 versus MBNL-AB, †P < 0.0001 versus MBNL-AB, Student's t-test).
Figure 3.
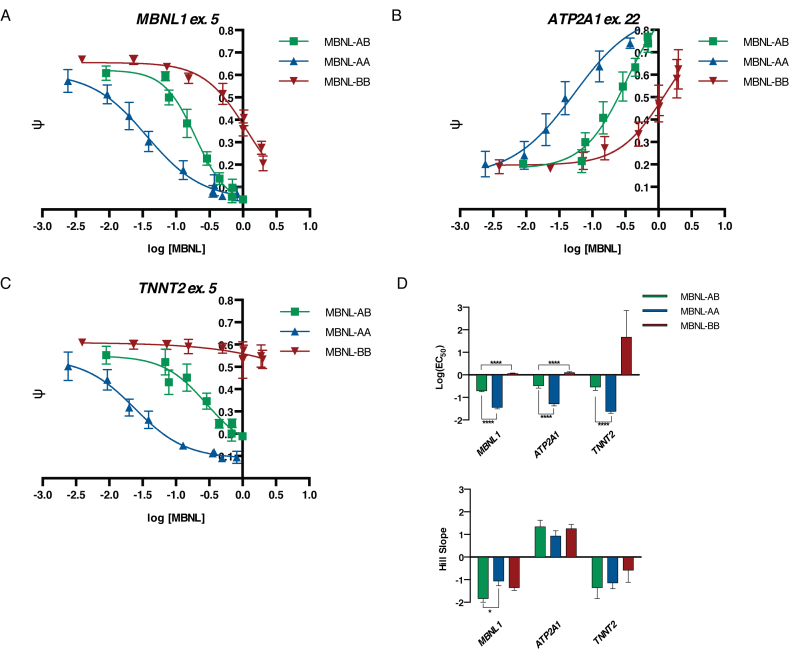
MBNL-AA and MBNL-BB proteins regulate splicing at different relative protein levels compared to MBNL-AB in a plasmid dosing system. (A–C) Plasmid dosing assays for MBNL1, ATP2A1, and TNNT2 minigene events, respectively. Increasing amounts of plasmid expressing MBNL-AB, MBNL-AA, or MBNL-BB were transfected into HeLa cells along with a minigene reporter (n = 3–5 per plasmid dose). ψ values (data represented as mean ± standard deviation) were then quantified, plotted against log [MBNL] levels, and fit to a four-parameter dose–response curve. Relative MBNL expression levels for each protein were determined via immunoblot (n = 3) at each plasmid dose and normalized to GAPDH. MBNL-AB levels at the highest plasmid dose (200 ng) were then set equal to 1 and all other values for MBNL-AB, MBNL-AA, and MBNL-BB normalized. Representative immunoblots with quantification and splicing gels can be found in Supplemental Figure S6 and S7A, respectively. (D) Bar plots of log(EC50) values and Hill slopes derived from the dose–response curves (table of exact values ± standard error are listed in Supplemental Figure S7B). Due to ambiguous curve fitting of MBNL-BB, the bottom (MBNL1 and TNNT2) or top (ATP2A1) of the curve was constrained to match the average ψ value of MBNL-AB at the highest plasmid dose. (*P < 0.05, ****P < 0.0001, Student's t-test).
Figure 7.
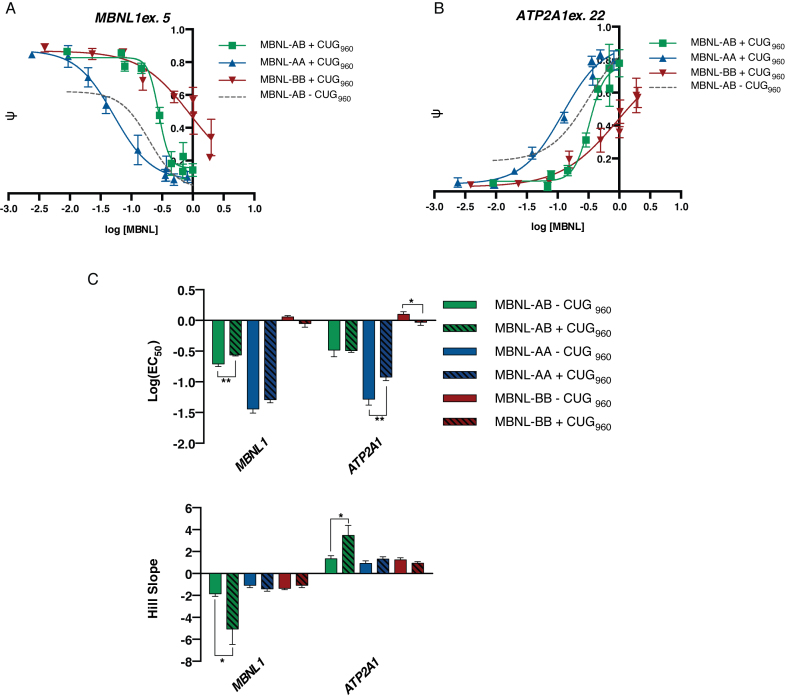
Dose curves of synthetic MBNLs are altered in the presence of toxic RNA expression. (A and B) Plasmid dosing assays were performed using the MBNL1 and ATP2A1 minigenes, respectively, in the presence of CUG repeat RNA expression. HeLa cells were transfected with the same increasing amounts of plasmid to create a gradient of protein expression as done in Figure 3. Cells were transfected with a minigene reporter and a CTG960 repeat expressing plasmid. ψ values at each plasmid dose were quantified (n = 3–4 for each plasmid dose), plotted against log [MBNL] levels, and fit to a four-parameter dose–response curve (data represented as mean ± standard deviation). Representative splicing gels can be found in Supplemental Figure S14. (C) Bar plots of log (EC50) values and slopes derived from the dose–response curves (table of exact values ± standard error are listed in Supplemental Figure S15C). Due to ambiguous curve fitting of MBNL-BB, the bottom (MBNL1) or top (ATP2A1) of the curve was constrained to match the average ψ value of MBNL-AB at the highest plasmid dose. These values are compared to those determined in the absence of toxic RNA expression (Figure 3D and Supplemental Figure S7B). Comparison of the dose curves in the presence and absence of CUG960 RNA expression can be found in Supplemental Figure S15A-B for MBNL1 and ATP2A1, respectively (*P < 0.05, **P < 0.01, Student's t-test).
MEFs were regularly maintained in DMEM-Glutamax supplemented with 10% FBS and 2 μg/ml puromycin at 37°C under 5% CO2. To assay endogenous splicing regulation (Figure 4B–D, Supplemental Figure S11) and GFP-MBNL expression levels via western blot (Figure 4A and Supplemental Figure S9), cells were plated in twelve-well plates at a density of 6 × 104 cells/well. After 24 h, fresh doxycycline was prepared at 1 mg/ml, diluted, and then added to the cells at the appropriate concentrations to induce a range of GFP-MBNL protein expression. 24 h post-docycycline treatment cells were harvested using TrypLE and pelleted using centrifugation.
Figure 4.
MBNL-AA regulates splicing at similar relative protein levels in an inducible tet-on system in mbnl 1/2 double knockout MEFs. (A) Representative immunoblots used to determine relative MBNL protein levels across a gradient of doxycycline treatment (0–60 ng/ml for MBNL-AB and 0–2000 ng/ml for MBNL-AA). Relative MBNL protein expression levels for each protein was determined via immunoblot (n = 3) at each doxycycline dose and normalized to GAPDH. MBNL-AB levels at the highest dose were then set equal to 1 and all other values for MBNL-AB and MBNL-AA normalized. Quantification of MBNL levels from triplicate immunoblots can be can be found in Supplemental Figure S9. (B–D) Dose curves of three endogenous splicing events Apbb2, Mta, and Depdc5, respectively. MEFs were treated with increasing amounts of doxycycline (n = 3) and ψ values quantified at each dose (representative splicing gels used to quantify ψ values can be found in Supplemental Figure S10). These ψ values (data represented as mean ± standard deviation) were then plotted against log [MBNL] levels and fit to a four-parameter dose-curve. Due to ambiguous curve fitting in some cases, for all dose–response curves, the top or bottom (i.e. inclusion or exclusion event, respectively) of the curve was constrained to match the average ψ at the highest doxycycline dose. Additional dose–response curves and R2 values for each curve fit can be found in Supplemental Figure S11. Quantitative parameters derived from these dose curves can be found in Supplemental Figure S12.
Western blot analysis
Cell pellets were lysed in RIPA (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) (ThermoFisher) supplemented with 1 mM phenylmethylsulfonyl fluoride and 1X protease inhibitor cocktail (SigmaFAST, Sigma) by light agitation for 15 min via vortex. Equal amounts of lysate were resolved on a 10% (HeLa/HEK-293 cells, Figure 1B and Supplemental Figure S3A) or 4–15% (MEF cells, Figure 4A) SDS-PAGE gel prior to transfer. For blots with lysates from HeLa/HEK-293 cells, MBNL proteins were probed using a mouse anti-HA antibody (1:1000 dilution, 6E2, Cell Signaling Technology) and goat anti-mouse secondary IRDye 800CW (1:15,000 dilution, LI-COR). A GAPDH loading control was probed using rabbit anti-GAPDH antibody (1:1000 dilution, 14C10, Cell Signaling Technology) followed by a goat anti-rabbit secondary IRDye 680RD (1:15,000 dilution, LI-COR). Blots from MEF lysates were probed with a rabbit anti-GFP (1:1000 dilution, D5.1, Cell Signaling Technology) and a donkey anti-rabbit secondary IRDye 680RD (1:15,000 dilution, LI-COR). A GAPDH loading control was probed using a chicken anti-GAPDH antibody (1:2000 dilution, ab14247, Abcam) followed by a donkey anti-chicken secondary 800CW (1:15,000 dilution, LI-COR). In both systems, fluorescence was measured using a LI-COR Odyssey Fc or LI-COR Odyssey CLx Imaging instrument. Quantification was performed using the associated Image Studio analysis software (LI-COR).
Cell-based splicing assay
RNA was isolated from HeLa and HEK-293 cells using an RNeasy kit (Qiagen) or Aurum Total RNA Mini kit (Bio-Rad). The isolated RNA was processed via reverse-transcription (RT)-PCR and the percent spliced in (PSI, ψ) (i.e. percent exon inclusion) for each minigene event upon protein or mock treatment was determined as previously described (31) (Figures 2–4 and 7 and Supplemental Figures S4, S11, and S15). The only differences from this previously published protocol was that for some RT steps SuperScript IV (Invitrogen) was utilized. Additionally, some cDNA samples were visualized and the percent exon inclusion (ψ) values determined using the Fragment Analyzer (DNF-905 dsDNA 905 reagent kit, 1–500bp, Advanced Analytical Technologies) and associated ProSize data analysis software. No discernible differences in ψ quantification was observed between samples visualized using 6% native gels and SYBR green I nucleic acid stain (Invitrogen) and the Fragment Analyzer system. In the plasmid dosing system with and without CUG repeat RNA expression (Figures 3 and 7), ψ values were plotted against relative MBNL levels as determined by immunoblot (Supplemental Figure S6) and fit to a four-parameter dose-curve (ψ = ψmin + ((ψmax - ψmin)/(1 + 10((log(EC50) – log[MBNL1]) * slope)))). Parameters that correlate to biological data, i.e. concentration (EC50) and steepness of response (slope), were then derived from these curves (Figures 3D and 7C).
RNA was isolated from MEF cells using the Aurum Total RNA mini kit and DNase treated on column. 1000 ng of DNAsed RNA was reverse transcribed with SuperScript IV with random hexamer priming according to the manufacturer's protocol except that half of the recommended SuperScript IV was utilized. cDNA was then PCR amplified for 25–32 cycles using flanking exon-specific primers. Primer sequences, annealing temperatures, and inclusion and exclusion product sizes in base pairs are listed in Supplemental Table S1. Samples were visualized and quantified using the Fragment Analyzer system. Ψ values were plotted against MBNL levels (Figure 4B–D, Supplemental Figure S11) as determined by Western plot (Figure 4A and Supplemental Figure S9) relative to GAPDH and fit to a four-parameter dose-curve as described above. Parameters that correlate to biological data, i.e. concentration (EC50) and steepness of response (slope), were then derived from these curves (Supplemental Figure S12).
Protein expression and purification
All proteins were expressed as N-terminal glutathione S-transferase (GST) fusions. Using BL21 Star (DE3) cells (Invitrogen), protein expression was induced using 0.5 mM IPTG at an OD600 = 0.6–0.7 for 2 h at 37°C. Following induction, cells were lysed in B-PER (bacterial protein extraction reagent) (Pierce) supplemented with DNase I (5 U/ml) and lysozyme (0.1 mg/ml) for 30 minutes at room temperature. The lysate was then diluted with 1 volume of 1× PBS and incubated for 30 minutes on ice prior to centrifugation at 17,000 rpm. The supernatant was isolated and mixed with glutathione agarose (Sigma) for 2 h at 4°C. The resin was washed twice with 5 volumes of GST buffer (40 mM bicine pH 8.3, 50 mM NaCl), twice with 5 volumes of GST buffer supplemented with 1 M NaCl, and finally 3 times with 5 volumes of GST buffer – 20 mM NaCl. GST-tagged MBNL-AB, MBNL-AA, and MBNL-BB were then eluted with 10 mM glutathione in GST buffer – 20 mM NaCl. The resulting elution was then concentrated and dialyzed into storage buffer (25 mM Tris pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol (β-ME), 50% glycerol). Final purity of the proteins was assessed via SDS-PAGE gel analysis and no significant differences were detected. The GST-MBNL fusions were the most prominent band with very few non-specific carryover products from the purification. Working concentrations were determined via the Pierce 660 nM protein assay reagent using BSA standards.
RNA radiolabeling and electrophoretic mobility shift assays (EMSAs)
All RNA substrates were ordered from IDT or Dharmacon and 5′ end-labeled using T4 PNK (NEB) with [γ-32P] ATP. All RNAs were purified on 10% polyacrylamide denaturing gels. Prior to incubation with protein, these RNAs were denatured by incubation at 95°C for 2 min followed by a 5 min incubation on ice. Once cooled the RNA was mixed with increasing concentrations of protein (final volume = 10 μl) to yield final reaction conditions of 115 mM NaCl, 20 mM Tris pH 7.5, 1 mM β-ME, 0.01 mM EDTA, 10% glycerol, 5 mM MgCl2, 0.1 mg/ml heparin, 2 mg/ml bovine serum albumin (BSA), and 0.02% xylene cyanol. This protein–RNA mixture was incubated for 30 minutes at room temperature for binding to reach equilibrium prior to electrophoresis. 3 μl of the sample was then loaded on a pre-chilled, 1.5 mm, 6% native acrylamide (37.5:1) gel and run for 45 min at 150 V at 4°C. Gels were dried for overnight exposure on phosphorus plates (Figure 4B and D). Binding curves were quantified using ImageQuant software (GE Healthcare Life Sciences). The fraction of RNA bound was calculated as the ratio of all RNA-protein complexes divided by total RNA signal in each lane. The apparent Kd was then determined using the following equation: fbound = fmax([MBNL]/([MBNL] + Kd)) (Figure 5C and E).
Figure 5.
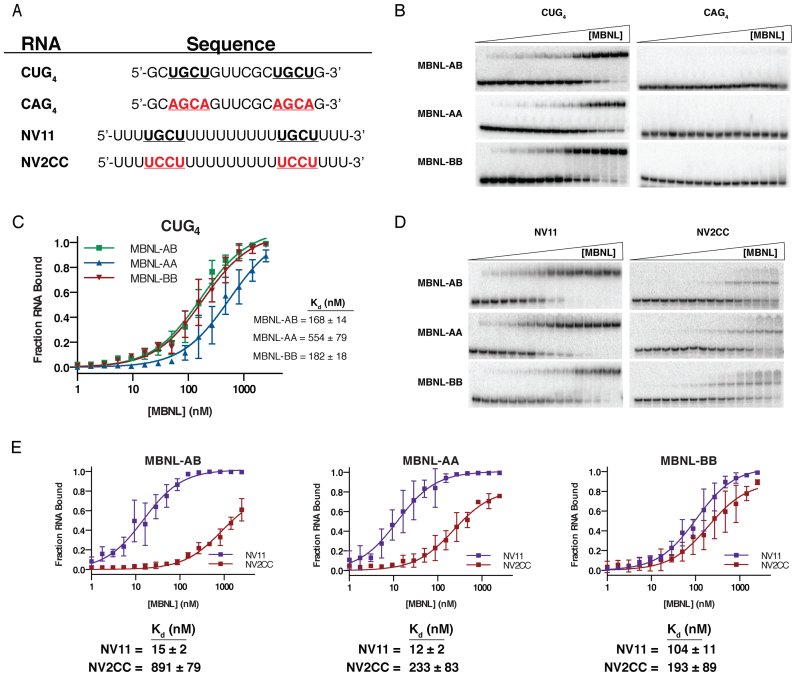
Reorganization of zinc finger domains does not significantly impact RNA binding of synthetic MBNL proteins. (A) Sequence of four RNAs used in EMSAs with synthetic MBNL proteins. The occurrence of specific UGCU motifs (bold and underlined) and mutated non-specific motifs (red, bold, and underlined) within the RNA substrates are noted. (B) Representative EMSA gels to CUG4/CAG4 RNA substrates (n = 3 for each RNA). (C) Binding curves of all synthetic MBNL proteins for CUG4. Apparent dissociation constants (Kds) (± standard error) for each MBNL1 are listed. (D) Representative EMSA gels for NV11/NV2CC RNA substrates (n = 3 for each RNA). (E) Binding curves for each MBNL protein comparing the differences in affinity between NV11 and the non-specific mutant NV2CC. Kd (± standard error) for each RNA are listed below each plot.
In vitro transcription of RNA Bind-n-seq (RBNS) random input RNA
RBNS random input RNA was prepared by in vitro transcription using the RBNS T7 template (5′-ACACTC TTTCCCTACACGACGCTCTTCCGATCT(N)40GATCGGAAGAGCACACGTCTGAACTCCAGTCACCCTATAGTGAGTCGTATTA-3′), a DNA oligo containing a random 40mer sequence flanked by priming sites for the addition of Illumina adaptors and the T7 promoter sequence. To artificially create a double-stranded T7 promoter, a T7 oligo (5′-TAATACGACTCACTATAGGG-3′) was annealed to the region of the RBNS T7 template corresponding to the T7 promoter sequence by heating the template and T7 oligo in equal proportions up to 95°C and cooling down at a rate of 0.1°C/s to 45°C. The RBNS input RNA pool was then in vitro transcribed using the HiScribe T7 in vitro transcription kit (NEB). The produced RNA was then bead purified using AMPure XP RNase free beads (Beckman Coulter Inc.).
RBNS and computational analysis
RBNS was performed using the same proteins purified as GST fusions for EMSAs. Eight concentrations of each MBNL protein (nM = 0,16, 32,125, 250, 500, 1000, 2000), including a no MBNL condition, were equilibrated in binding buffer (25 mM Tris pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.01% Tween-20, 1 mg/ml BSA, 1 mM DTT, 30 μg/ml poly I/C (Sigma)) for 30 min at room temperature. In vitro transcribed RBNS random input RNA was then added to a final concentration of 1 μM with 40 U of SUPERaseIn (Ambion) and incubated for 1 h at room temperature. During this incubation 50 μl aliquots of glutathione magnetic agarose beads (Pierce) were washed four times with 0.2 ml of wash buffer (25 mM Tris pH 7.5, 150 mM KCl, 60 μg/ml BSA, 0.5 mM EDTA, 0.01% Tween-20). The beads were then placed in 50 μl of binding buffer until needed. To pull down the tagged MBNL and interacting RNA, each RNA and protein solution was added to 15 μl of equilibrated and washed glutathione magnetic agarose beads and incubated for 1 hour at room temperature. Unbound RNA was removed by washing the beads three times with 0.2 ml of wash buffer. The beads were incubated at 70°C for 10 min in 100 μl of elution buffer (10 mM Tris pH 7.0, 1mM EDTA, 1% SDS) and the eluted material (bound RNA) collected with AMPure XP RNase-free beads. The RNA was then reverse transcribed into cDNA using SuperScript IV according to the manufacturer's protocol with a common primer (5′-ACTGACCTCAAGTCTGCACACGAGAAGGCTAG-3′). 0.5 pmol of RBNS input RNA was also reverse transcribed to control for any nucleotide biases in the input library. Illumina sequencing libraries were prepared using primers with Illumina adaptors and unique sequencing barcodes (to allow for multiplexing all samples) to amplify the cDNA using Phusion high-fidelity DNA polymerase (NEB) for 16 amplification cycles. Table of primers used to index each sample library with unique barcodes are listed in Supplemental Table S2. PCR products were bead purified using AMPure XP RNase-free beads. Sequencing libraries corresponding to all concentrations of a given MBNL were pooled in a single lane and the random 40mer sequenced using the Illumina NextSeq 500. Motif (kmer) R values were calculated as the motif frequency in the selected RBP pool over the frequency in the input RNA library (Figure 6A–C). Frequencies were controlled for the respective library read depth. The overall rate of kmer enrichment in the no protein condition relative to the input library was defined as the false-discovery rate (FDR). More detailed methods and theoretical assumptions utilized have been previously reported (36).
Figure 6.
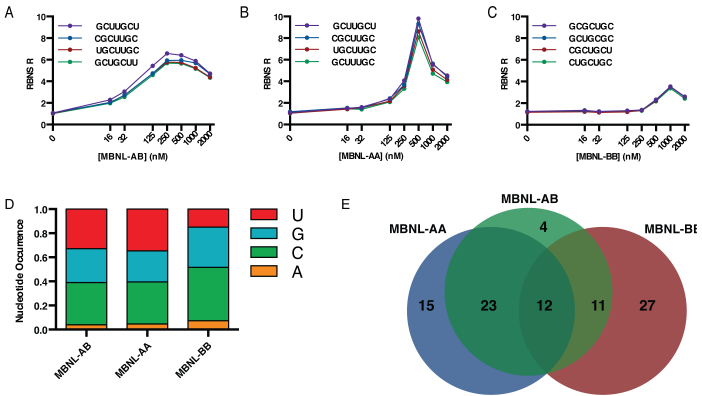
RBNS analysis of engineered MBNL proteins indicates that the ZF domains have differential RNA binding specificity. (A–C) RBNS R values for the top four kmers (k = 7) are shown as a function of MBNL1 protein concentration for MBNL-AB, MBNL-AA, and MBNL-BB, respectively. Top four kmers for each protein are determined based on concentration of protein that shows the greatest R values (250, 500 and 1000 nM for MBNL-AB, MBNL-AA, and MBNL-BB, respectively). R values at all other concentrations for the respective kmers were then determined to create the unimodal enrichment plots shown. (D) Percent nucleotide occurrence within the top 100 kmers for each MBNL protein. (E) Area-proportional Venn diagram showing overlap in top 50 kmers for each MBNL protein. Values listed represent number of kmers within each sub-population. [RBNS data can be accessed via Sequence Read Archive SUB2513163; RNA Bind-N-Seq for Synthetic MBNL.]
RESULTS
Synthetic MBNL1 proteins with modified zinc finger domain organization possess different splicing activities
In order to evaluate the importance of ZF domain organization and content in MBNL proteins, two synthetic MBNL proteins with different ZF domain content were created, MBNL-AA and MBNL-BB, the activities of which we planned to compare to an MBNL protein with the canonical ZF domain content and arrangement, MBNL-AB (Figure 1A). Using the extensively studied 41 kDa isoform (1–382 amino acids) of MBNL1 as a platform for our synthetic protein design, we defined the ZF1–2 domain (domain A) from 9 to 101 (93 amino acids) and the ZF3–4 domain (domain B) from 178 to 253 (76 amino acids). Although the domain boundaries previously published were used as a guide (33,44), we chose to extend the C-terminal sequence of ZF1–2 to include the Q-rich region (amino acids 91–101) downstream of ZF2, which we have shown previously to be important for ZF1–2 splicing function (31). To reduce the overall size of our synthetic proteins and facilitate in vitro purification, the C-terminal region (amino acids 261–382) was removed and replaced with an eight amino acid nuclear localization signal (SV40 NLS). Although predicted to be relatively unstructured, the C-terminus of MBNL1 has been shown to contain several regions required for nuclear localization and potential MBNL1 dimerization (45). However, previous work has shown that this region is not required for high-affinity RNA binding and MBNL1 proteins with the C-terminus removed retain nearly full splicing activity compared to full-length MBNL1 (41,46). Finally, a N-terminal HA tag was also added for use in immunoblot and immunofluorescence detection methods (see Supplemental Figure S1A for the amino acid sequences of all MBNL protein constructs used in this study).
Prior to functional characterization of our synthetic MBNL proteins, we evaluated relative protein expression levels and subcellular localization. Immunofluorescence detection in transfected HeLa cells showed predominant nuclear localization with a modest signal in the cytoplasm for MBNL-AB and both synthetic proteins (Supplemental Figure S2A). This distribution is comparable to past results, including those using full-length MBNL1 (31,39). The only noticeable difference in the subcellular distribution of the synthetic proteins was a lack of nucleolar definition in cells expressing MBNL-BB. Surprisingly, we detected significant differences in steady state protein levels in transfected HeLa cells as determined by immunoblot (Figure 1B). When normalized to MBNL-AB, MBNL-AA is expressed at an approximately 0.5-fold lower level while MBNL-BB is expressed at a 2.5-fold higher level (Figure 1C). This pattern of expression was maintained in transfected HEK-293 cells indicating that the observed relative expression levels observed are independent of cell-type and transfection method (Supplemental Figure S3A-B). These variations in protein levels were not due to changes in mRNA expression as assayed by RT-qPCR (Supplemental Figure S2B). Overall, these data suggest that the ZF3–4 domain confers additional stability to MBNL1 compared to ZF1–2 in the context of these protein constructs.
To explore how variation of ZF content within the synthetic proteins would impact MBNL1 splicing activity, a cell-based splicing assay was used with a series of splicing reporter minigenes, many of which are derived from events known to be mis-regulated in DM. These reporters include (i) human insulin receptor exon 11 (INSR) (44,47,48), (ii) human cardiac troponin T type 2 exon 5 (TNNT2) (35,41), (iii) human sarcoplasmic / endoplasmic reticulum Ca2+-ATPase 1 exon 22 (ATP2A1) (32,49), (iv) mouse nuclear factor I/X exon 8 (Nfix) (17), (v) mouse very-low-density lipoprotein receptor exon 16 (Vldlr) (17), and (vi) human MBNL1 exon 5 (50). HeLa cells were co-transfected with synthetic MBNL-expression plasmids or empty vector (mock) and a single minigene reporter. Inclusion levels of each alternative exon were then quantified via RT-PCR and expressed as percent spliced in (PSI, ψ) (i.e percent exon inclusion). Splicing activity of MBNL-AA and MBNL-BB were then determined as a percentage of activity relative to MBNL-AB for each minigene event.
The data for the six minigenes tested revealed that MBNL-AA regulated splicing at a level equivalent to or better than MBNL-AB. MBNL-BB, while still functional, had significantly reduced splicing activity (Figure 2A–F). These patterns of splicing regulation were maintained for both inclusion (INSR, ATP2A1 and Vldlr) and exclusion (TNNT2, MBNL1 and Nfix) events, indicating that the splicing activity of these synthetic proteins is independent of RNA target and regulation type. Overall, these observations are consistent with our hypothesis that the splicing activity of MBNL-AA would be high while that of MBNL-BB would be low. Importantly, disruption of the canonical ZF domain organization and removal / replacement of specific ZF domains did not render our synthetic MBNL proteins dysfunctional, indicating that (i) MBNL proteins are amenable to major sequence alterations and substitutions, and (ii) the splicing activity of the individual ZF domains can be uncoupled.
The only reporter that showed large differences in regulation was TNNT2. Within the context of this event, MBNL-AA displayed enhanced activity (147%) while MBNL-BB had only minimal splicing activity (16% activity) (Figure 2D). In contrast, all three proteins were able to regulate splicing of the MBNL1 reporter with similar activity (Figure 2E). For all other reporters utilized, MBNL-AA regulated splicing at equivalent levels to MBNL-AB while MBNL-BB retained ∼50% of MBNL-AB splicing activity (Supplemental Figure S5A). These trends were maintained in HEK-293 cells despite changes in Δψ for each minigene between the two different cell types (Supplemental Figures S4A-F and S5B).
An important point regarding these results is that equal amounts of plasmid were transfected into cells and this resulted in differences in the amount of each MBNL protein expressed (Figure 1B–C for HeLa, Supplemental Figure S3 for HEK-293). Interestingly, the high levels of MBNL-BB were not sufficient to regulate splicing as well as MBNL-AB. In contrast, MBNL-AA maintained comparable splicing regulation to MBNL-AB with half the amount of protein present.
Controlled dosing of synthetic MBNL proteins in two different systems reveals significantly different activities for splicing regulation
To gain further insight into MBNL-AA and MBNL-BB AS regulation, especially as it relates to protein concentration, we performed the same cell-based splicing assays previously utilized across a gradient of MBNL expression. We found that this experimental analysis was necessary as our synthetic proteins had different expression profiles (Figure 1B–C, Supplemental Figure S3). To create the range of protein levels required within this system, HeLa cells were transfected with increasing amounts of protein-expression plasmid for each synthetic MBNL protein tested. Immunoblot analysis against the HA tag was then used to quantify relative MBNL1 levels at each concentration of plasmid transfected (see Supplemental Figure S6 for representative blots and quantification). As expected, MBNL-BB maintained relatively high levels of expression across the gradient while MBNL-AA protein levels remained lower compared to MBNL-AB.
Next, ψ values for three different minigenes (TNNT2, MBNL1 and ATP2A1) for each individual point along the protein gradient were determined (representative images used to calculate ψ are shown in Supplemental Figure S7A). These values were then plotted against log [MBNL] to create dose–response curves for each protein. MBNL1 and ATP2A1 were selected from the pool of mingene reporters to test in this system because (i) these two minigenes displayed a robust splicing response (large Δψ) in the cell-based splicing assay (Figure 2B and E), (ii) they represent both MBNL1-regulated inclusion and exclusion events, respectively, (iii) both minigenes have been well-characterized (32,50) and (iv) MBNL-AA and MBNL-BB both show similar splicing activity and maximal ψ compared to MBNL-AB (Figure 2B and E). TNNT2 was chosen as an additional reporter to test in this dosing system because it displayed the largest difference in splicing activity between the synthetic MBNL proteins (Figure 2D). Creation of these dose curves allowed for the derivation of several quantitative parameters that describe the splicing regulation of each event, i.e. EC50 and slope. The slope of the response curve provides a relative measure of cooperativity while the EC50 value provides a relative measure of how much protein is required to obtain splicing regulation at 50% of maximum ψ.
Results from these experiments revealed different dose–response curves for each MBNL protein tested and for each minigene assayed (Figure 3A–C). Both MBNL-AB and MBNL-AA displayed typical dose response curves that show a plateau in ψ for all three minigene events tested (Figure 3A–C). Based on the EC50 values derived, MBNL-AB required ∼5-fold more protein compared to MBNL-AA to achieve similar levels of splicing regulation (Figure 3D). For all three events tested the slope of the dose response curves for MBNL-AB was steeper compared to MBNL-AA (Figure 3D). Interestingly, this indicates that while less MBNL-AA protein is required to reach the maximum ψ, there is an apparent loss in cooperative splicing regulation. In contrast to MBNL-AB and MBNL-AA, the dose–response curves for MBNL-BB revealed that, as expected, high expression levels are required to achieve modest splicing regulation (Figure 3D). Almost no change in ψ was observed for TNNT2 (Figure 3C). Even for minigene events assayed in which MBNL-BB was able to achieve splicing regulation in the overexpression system (ATP2A1 and MBNL1, Figure 2B and E), the EC50 values are high and the slopes are shallow compared to the other two proteins (Figure 3D). Overall, the controlled dosing of our synthetic MBNL1 proteins in this system revealed that as predicted, MBNL-BB has significantly reduced splicing activity while MBNL-AA should be considered a high activity, synthetic derivative of MBNL-AB with a 5-fold increase in splicing activity.
To expand our analysis of synthetic MBNL AS regulation as a function of protein concentration, we established stable cell lines expressing GFP-tagged MBNL-AB or MBNL-AA controlled with tet-on regulation. A cell line with MBNL-BB was not generated due to its weak splicing activity. Both a constitutively expressed rtTA and an N-terminal GFP-tagged synthetic MBNL protein under control of a tet-response element were stably integrated into mbnl 1/2 double knockout mouse embryonic fibroblasts (MEFs). Integration of both cassettes was driven by puromycin selection. After selection and treatment with doxycycline to activate GFP-MBNL protein expression, fluorescent-activated cell sorting was used to isolate and select individual clones for each cell line that have high expression of the synthetic MBNL protein in response to drug treatment. The fluorescence of the GFP tag was utilized to show that as in the transfected HeLa system, MBNL-AB and MBNL-AA co-localize in the nucleus of doxycycline treated cells (Supplemental Figure S8).
In this system, the concentration of synthetic MBNL proteins can be precisely controlled as a function of doxycycline (0–60 ng/ml for MBNL-AB, 0–2000 ng/ml for MNBL-AA). In both cell lines, MBNL expression covered a broad range (Figure 4A, Supplemental Figure S9). In contrast to the plasmid dosing system in HeLa cells, expression levels of MBNL-AB and MBNL-AA at matched doxycycline doses are statistically equivalent except at the highest dose, where MBNL-AA expression levels were slightly increased (Supplemental Figure S9). The differences in protein expression levels in the two systems are likely due to the presence of the N-terminal tag (GFP versus HA) and possibly the different cellular environments. Next, we tested the AS activity of the synthetic MBNL proteins for 15 endogenous splicing events across a range of protein expression generated via doxycycline gradient. These 15 endogenous events (9 inclusion, 6 exclusion) were selected from RNAseq data sets previously published from the mbnl 1/2 knockout MEFs (39). RT-PCR was then performed to determine ψ of each individual point along the protein gradient (representative images to calculate ψ are shown in Supplemental Figure S10) and plotted against log [MBNL] levels to generate dose–response curves (Figure 4A–C, Supplemental Figure S11). EC50, slope, and Δψ values were then derived from these dose–response curves (Supplemental Figure S12).
MBNL-AB and MBNL-AA displayed nearly identical dose–response curves with similar EC50 and slope values (Figure 4B–D and Supplemental Figure S11). In most cases, the dose–response curves overlapped (Figure 4B, Supplemental Figure S11C-G, S11J, and S11L). For a few select events, while the overall shape / Δ ψ of the dose–response curves was similar, the minimal and maximal ψ for MBNL-AB or MBNL-BB was shifted (Figure 4C and Supplemental Figure S11B, S11E, S11I and S11K). These shifts in the curves did, for some events (Mta, Add3, and Exoc1) result in increased EC50 values for MBNL-AA compared to MBNL-AB (Supplemental Figure S12A-B). Depdc5 (Figure 4D) was the only event for which MBNL-AA showed a significantly lower EC50 and reduced slope compared to MBNL-AB, the same pattern of activity displayed in the HeLa plasmid dosing system. Overall, MBNL-AB and MBNL-AA showed similar activities across many splicing events suggesting these two proteins regulate most or all splicing events with similar activities within the context of this system.
Synthetic MBNL proteins possess distinct RNA binding specificities
To determine if enhanced or disrupted RNA binding correlated with the observed splicing activities of MBNL-AA and MBNL-BB, electrophoretic mobility shift assays (EMSAs) were performed with purified MBNL proteins and short model RNAs. The first tested was a CUG4 RNA substrate which contains two UGCU motifs (Figure 5A) predicted to form a short hairpin designed to mimic the structure CUG repeats are proposed to adopt in DM1 (41). Surprisingly, MBNL-AB and MBNL-BB possessed nearly identical binding affinities to the CUG4 RNA while MBNL-AA had a slightly higher KD (Figure 5B). As expected, all three proteins had no observable binding to the CAG4 RNA substrate (Figure 5A) in which the UGCU motifs were mutated to AGCA to weaken MBNL1-RNA interactions (Figure 5B) (all Kds > 2500 nM).
Second, we assayed binding to NV11, a 24-nucleotide, single-stranded RNA substrate that serves as a model for sites in pre-mRNAs with minimal RNA structure (34). This RNA contains two GC dinucleotides separated by an eleven uridine spacer creating two UGCU binding motifs (Figure 5A) (34). In conjunction we tested the NV2CC substrate in which both GC dinucleotides are mutated to CC (Figure 5A). This modification to the sequence leads to disruption of the YGCY binding motifs (UGCU to UCCU) and has been shown to significantly weaken MBNL1 binding (34). MBNL-AB and MBNL-AA had nearly identical, low nanomolar binding affinities to the NV11 substrate (Figure 5D–E). In a manner similar to CAG4, both proteins displayed a substantial decrease in RNA binding affinity to the NV2CC construct (Figure 5D–E). Differential protein–RNA complex migration was observed for NV2CC compared to NV11. We suggest these differences are due to alterations in the binding mode of the MBNL proteins for specific vs. non-specific binding motifs. Overall, these results indicate that MBNL-AB and MBNL-AA both recognize YGCY motifs with relatively high levels of specificity (59-fold increased recognition of specific motifs for MBNL-AB and 18-fold for MBNL-AA).
MBNL-BB exhibited a 6-fold decrease in RNA binding affinity for the NV11 RNA substrate compared to MBNL-AB (Figure 5D–E). Interestingly, MBNL-BB only displayed a 2-fold decrease in RNA binding affinity for NV2CC compared to NV11 (Figure 5D–E). This result indicates that MBNL-BB partially lost the ability to specifically recognize target YGCY motifs in the context of a pyrimidine rich RNA. This pattern is significantly different from both MBNL-AA and MBNL-AB, as both proteins exhibit high affinities for NV11 with significantly increased KDs for NV2CC (Figure 5D–E). Overall, these data suggest that MBNL-BB is primarily a non-specific RNA binding protein. Although we originally predicted that the differences in splicing activity observed between the synthetic MBNL1 proteins would be due to changes in RNA binding affinity, the results from our EMSA analysis suggested that differences in RNA binding specificity might be responsible.
To more broadly test the RNA binding specificities of the synthetic MBNL proteins, we performed RNA Bind-n-Seq (RBNS), a comprehensive, next-gen sequencing based approach to characterize sequence specificity of RBPs (36). MBNL-AB, MBNL-AA and MBNL-BB were incubated at increasing concentrations with a pool of random 40-mer RNAs. The bound RNA, as well as a sample of the un-processed input RNA, was then used to produce cDNA libraries for deep sequencing (36). Following sequencing, for each protein at each of the tested concentrations, motif read enrichment, or ‘R’ values, were calculated for each kmer (k = 7) as the ratio of the frequency of the kmer in the experimental pool as compared to that of the input RNA library. Using this approach, a higher R value is indicative of increased enrichment of a specific motif in the bound RNA pool where R = 1 indicates no significant enrichment.
First, we compared data from our RBNS analysis of MBNL-AB to that of a previously published RBNS MBNL1 data set (36). We observed many of the same top kmers in both RBNS analyses as well as similar R-values with correlations across the range of protein concentrations (Supplemental Figure S13B-C). This indicates that (i) our MBNL-AB protein had similar levels of binding activity compared to the truncated MBNL1 protein utilized in other independent studies and (ii) there is only a modest difference between the experiments likely due to the use of different tags in the experimental methodology and changes in the washing step of the protocol (GST versus a streptavidin binding peptide (36), see Materials and Methods for additional information about experimental design).
Next, we compared the unimodal enrichment profiles of the top four kmers for MBNL-AB and the two synthetic MBNL derivatives (Figure 6A–C). Analysis of these plots revealed several interesting patterns. First, the top three kmers identified were the same for MBNL-AB and MBNL-AA (GCUUGCU, CGCUUGC and UGCUUGC). All three kmers contain either YGCU or GCUU motifs, with the top kmer of GCUUGCU containing both motifs. Overall, there was significant overlap in the top 50 kmers identified for both proteins as well as similar nucleotide occurrence in the selected motifs (Figure 6D–E). This indicates that both MBNL proteins recognize and bind similar RNA motifs.
Interestingly, the R values for many top kmers are significantly increased for MBNL-AA compared to MBNL-AB (10 versus 7), albeit at different protein concentrations (500 nM vs 250 nM) (Figure 6B and A). Although this is the most striking difference between these two proteins, the overall pattern is that at lower concentrations, MBNL-AA has lower R values compared to MBNL-AB until these R values dramatically increase at 500 nM, and then drop to nearly identical levels at higher protein concentrations (Figure 6B). In contrast, R values for MBNL-AB increase modestly at lower concentrations, peaking at 250nM and then staying relatively constant (Figure 6A). This overall pattern suggests that higher concentrations of MBNL-AA may be needed to achieve specific sequence binding relative to MBNL-AB, potentially due to loss of cooperative binding as was suggested by the shallower slopes of the splicing dose–response curves generated using minigene reporter substrates (assuming RNA binding correlates to splicing). Despite changes in the shape of the enrichment profiles, there is high correlation in the R values across the protein gradient (Supplemental Figure S13A). Overall, RBNS analysis indicates that while MBNL-AA and MBNL-AB bind and recognize similar RNA motifs, MBNL-AA has increased RNA binding specificity for many of these motifs.
MBNL-BB selected a different set of top kmers that contain fewer uridines (GCGCUGC, GCUGCGC, CGCUGCU, and CUGCUGC) (Figure 6C). Percent nucleotide occurrence within the top 100 kmers showed a reduction of uridines and a modest enrichment in guanosines and cytosines (Figure 6D). Due to this change in the distribution of nucleotides in the enriched MBNL-BB kmers, fewer YGCU and UGCU motifs were identified. As such, fewer overlapping motifs were found between the top 50 kmers of MBNL-BB and the other MBNL proteins (Figure 6E). Consistent with modest RNA binding specificity, the enrichment profiles for MBNL-BB have low R values across the gradient of protein concentrations, peaking at R = 3 at 1000 nM for the most enriched motifs (Figure 6C). Overall, the EMSA and RBNS data for MBNL-BB indicate that this synthetic MBNL1 has significantly reduced RNA binding specificity while maintaining general RNA binding affinity. This is in sharp contrast to MBNL-AA in which RBNS revealed that this synthetic protein has enhanced YGCY RNA sequence recognition over MBNL-AB. These overall changes in binding specificity are consistent with a model in which ZF1–2 confers specific sequence recognition while ZF3–4 acts as a more general RNA binding domain in the context of MBNL proteins and our synthetic derivatives.
Synthetic MBNL proteins rescue CUG-dependent mis-splicing in a DM1 cell model
Given the differences in splicing activity and RNA binding specificity of the synthetic MBNL proteins, we sought to determine if these proteins could rescue CUG-mediated mis-splicing like that found in DM1. This was accomplished by expressing CUG repeats from a plasmid containing 960 interrupted CTG repeats (DMPK-CTG960, a.k.a. CUG960) in culture (51). Interrupted repeats were used due to challenges with the instability of pure repeats (52) and within previous studies, the use of these long repeats with interruptions in HeLa cells leads to MBNL co-localization with CUG repeat RNA in nuclear foci and mis-splicing of MBNL-regulated minigenes (32,41,50). The same plasmid dosing system used previously (Figure 3) was used with co-transfection of the CUG960 repeat plasmid, minigene reporter, and synthetic MBNL expression construct to monitor ψ changes across the gradient of protein expression for the MBNL1 and ATP2A1 minigenes (representative images used to calculate ψ shown in Supplemental Figure S14). These results were also compared to those generated in the absence of CUG repeat RNA expression (Supplemental Figure S15).
Co-expression of CUG960 led to reduced splicing activity for all three proteins at low protein levels (Figure 7A–B), presumably due to sequestration of endogenous MBNL proteins. At higher protein expression levels, all three MBNL proteins were able to reach maximal splicing regulation (maximal ψ) equivalent to that in the absence of CUG repeats (Figure 7A–B, Supplemental Figure S15A-B). The addition of CUG repeats had no effect or only a modest effect on the EC50 values for all three proteins for both minigene reporters (Figure 7C). An increase in the slopes of the dose–response curves for MBNL-AB for both events studied was significant (Figure 7C). The overall effects on the dose–response curves with the addition of CUG repeats are consistent with a model in which at low levels of MBNL all of the protein is sequestered by the CUG repeats and unable to regulate splicing. As the concentration of MBNL increases binding to the CUG repeats is saturated, leading to a replenishment of free, active MBNL in the nucleus and effective splicing rescue. Despite the changes in the dose–response curves in the presence of toxic RNA, MBNL-AA remained the most active protein (lowest EC50 values) for both minigene reporters (Figure 7C).
DISCUSSION
Synthetic MBNL proteins with altered RNA binding specificity have differential splicing activity
To gain insight into the function of the individual ZF domains, we utilized a synthetic biology approach to engineer and biochemically characterize two synthetic MBNL proteins with altered ZF domain content, i.e. MBNL-AA and MBNL-BB. Using this system we determined that ZF1–2 has increased RNA binding specificity over ZF3–4 that led to enhanced AS activity of the synthetic MBNL-AA protein. Additionally, we showed that MBNL-AA was capable of rescuing CUG-dependent mis-splicing in a DM1 cell model at lower protein concentrations than MBNL-AB, indicating that these synthetic proteins could potentially be used as therapeutics to replace or displace sequestered MBNL from foci in DM patient cells.
Splicing assays, in vitro EMSAs, and RBNS analysis revealed that MBNL-AA is a more active derivative of MBNL1 and can regulate AS of RNA targets at reduced protein concentrations (Figures 3 and 7). Overall, MBNL-AA had either equivalent or 5-fold increased activity compared to MBNL-AB depending on the splicing assay and cell-system utilized. We predict that the differences in activity observed for MBNL-AA in the transfection experiments versus the inducible-expression system in the mbnl 1/2 double knockout MEFs is likely due to (i) differences in the N-terminal tag (HA versus GFP, respectively) and/or (ii) assessing AS with minigene events versus endogenously expressed pre-mRNA substrates in the two systems. Fusion of MBNL-AB and MBNL-AA to a large protein tag has the potential to alter the proteins’ overall activity and ability to load onto RNA substrates for effective regulation. Additionally, it is possible that the differences in MBNL-AB and MBNL-AA activity are magnified with RNA substrates at high cellular concentrations produced from transfected minigenes. Overall, these results indicate that protein modifications (tags), cellular environment and substrate concentrations can affect MBNL1 protein activity, but RNA binding specificity of MBNL1 is a primary determinant of its splicing regulation.
In contrast to MBNL-AA, MBNL-BB, while still functional, had 4-fold weaker activity compared to MBNL-AB. We predicted that these variations in splicing activity would be due to altered binding affinity to RNA targets, but we found through EMSA and RBNS studies that changes in RNA specificity appear to be primarily responsible for the observed alternations in splicing regulation (Figures 5 and 6). MBNL-AA retained the ability to recognize YGCY motifs with increased specificity compared to MBNL-AB (Figure 6). In contrast, MBNL-BB had very low RNA binding specificity overall with diminished recognition of canonical YGCY motifs (Figure 6).
Overall, our working model (summarized in Figure 8) is that in the context of the canonical arrangement of ZF domains in MBNL proteins (MBNL-AB), ZF1–2 drives splicing activity via specific binding to YGCY motifs in the appropriate sites of pre-mRNA substrates. ZF3–4, with its modest preference for YGCY motifs, will sample and bind many RNA motifs providing general binding affinity for MBNL. We propose that MBNL-AA with two high specificity domains has heightened recognition of MBNL1 YGCY regulatory elements leading to increased splicing activity. In contrast, our model is that MBNL-BB will bind many off-target sites leading to reduced occupancy at the sites needed for regulation of AS by MBNL1, resulting in the need for high concentrations of this protein for splicing regulation. In this model, the addition of a third ZF1–2 domain to create the synthetic protein MBNL-AAA would lead to enhanced interactions with multiple YGCY motifs and improved specificity of RNA recognition and regulation.
Figure 8.
Model summarizing differences between synthetic MBNL proteins. MBNL-AA is a more active alternative splicing regulator while MBNL-BB is significantly weaker compared to MBNL-AB. These differences in activity are represented by the size of the arrows showing how each MBNL protein promotes exon inclusion / exclusion. In the context of the canonical arrangement of ZF domains within MBNL proteins (MBNL-AB), ZF1–2 binds YGCY motifs with high specificity while ZF3–4 has a modest preference for YGCY motifs but will sample and bind many motifs with similar affinities. This activity is represented by the dotted lines illustrating ZF3–4 binding to both canonical and non-canonical RNA motifs. MBNL-AA possesses two high-specificity ZF1–2 motifs driving RNA recognition and subsequently increased splicing regulation at lower protein concentrations. MBNL-BB has significantly reduced RNA binding specificity and samples many specific and non-specific RNA motifs. Due to the reduced motif recognition, regulatory sites are not bound until high concentrations of protein are present leading to an overall reduction in splicing regulation. Structures of domains shown here are derived from PDB ID 3D2N (ZF1–2) and PDB 3D2Q (ZF3–4) (33).
The results from the cell-based splicing assays are consistent with our proposed model of AS regulation by the synthetic MBNL proteins. In general, MBNL-BB weakly regulated all tested minigene events, but achieved nearly complete splicing regulation with MBNL1 and ATP2A1. Both events possess many functional clusters of YGCY motifs (32,53), and MBNL-BB with its low specificity may bind the high density sites with sufficient occupancy to regulate splicing. The TNNT2 substrate contains only two UGCU motifs separated by the polypyrimidine tract within intron 4 (41), and these two sites may not be sufficient to recruit MBNL-BB to this substrate accounting for the acutely weak regulation of this event. Alternatively, it is possible that the synthetic MBNL proteins have altered recognition of specific RNA structural elements. Recognition of a structured element within the TNNT2 pre-mRNA and its subsequent disruption into a single-stranded segment is proposed to be required for regulation by MBNL1 (37,41). MBNL-AA may have enhanced recognition of structured RNAs, which may account in part for its increased splicing regulation of this event. An alternative possibility is that the ZF1–2 domain interacts with other splicing factors that bind the TNNT2 pre-mRNA and the duplication of the ZF1–2 domain improves recruitment and leads to the higher level of splicing activity observed (Figures 2D and 3C).
ZF1-2 and ZF3-4 possess distinct RNA binding activities that modulate MBNL1 activity
Previous work in the field had attempted to determine the activities and RNA binding of the individual ZF1–2 and ZF3–4 domains and how each contributes to the overall function of MBNL1 (31). Truncated MBNL1 proteins possessing only ZF1–2 or ZF3–4 bind weakly to RNA substrates, suggesting that the tandem ZFs work cooperatively to bind their RNA targets (54). Other studies have utilized point mutations to eliminate the RNA binding function of one ZF pair while leaving the other functional (31). It was shown using this strategy that ZF3–4 binds RNA with lower affinity than ZF1–2 (31). While these results are consistent with our binding studies and RBNS analysis, previous studies had not addressed questions of RNA binding specificity. The duplication of the ZFs in this study was important because it allowed us to study MBNL proteins that maintained nanomolar binding affinity for RNA targets and revealed significant differences in specificity. This duplication strategy should be useful for other domains that bind RNA with low affinity in isolation, assuming the domains operate independently.
Overall, the results with our synthetic MBNL proteins indicate that ZF1–2 and ZF3–4 are independent domains and can be reorganized without obvious negative impacts on protein function. Our studies indicate that ZF1–2 drives splicing regulation (Figures 2–4) via specific recognition of YGCY motifs (Figure 6). This activity is consistent with observations that MBNL1 orthologs in D. melanogaster and C. elegans which contain only a single ZF pair orthologous to ZF1–2 can regulate exon inclusion in a manner similar to human MBNL1 in mammalian cell culture (39). The differential protein levels of the synthetic proteins (Figure 1B–C, Supplemental Figure S3), suggest that the ZF3–4 domain may confer stability to MBNL1. Consistent with this hypothesis, increased mammalian and bacterial protein expression levels were observed for MBNL-BB; in contrast levels of MBNL-AA were reduced in both systems except for the inducible-MEF system where fusion to a GFP tag may stabilize relative protein levels. Interestingly, in many of the immunoblots performed in this study, two bands for MBNL-BB, and to a lesser extent, MBNL-AB, can be visualized. These bands could represent incorrectly processed protein or differential levels of post-translational modifications, which may in part account for the MBNL-BB’s reduced activity.
We propose that the differential activities between ZF1–2 and ZF3–4 activity observed in this study, namely the difference in RNA binding specificity and splicing activity, are due to subtle changes in the architecture and sequence of the ZF domains (Supplemental Figure S1B). Specifically, the N-terminal helical turn of ZF1 and the slightly extended C-terminal helix of ZF2 that are both absent in the ZF3–4 domain. Both of these structural elements were shown to be important for coordinating RNA in the binding pocket observed in the NMR solution structure of ZF1–2 bound to a short, human TNNT2 RNA fragment containing YGCY MBNL1 binding sites (37). We propose that the coordination and packing of RNA along the extended C-terminal helical element aids in specific-RNA recognition, making this region of the ZF act as a RNA discrimination domain. Although the structures of ZF1–2 and ZF3–4 are similar (33,37), due to the absence of the N-terminal helical turn in ZF3 and the shortened C-terminal helical region of ZF4 (33,37), we predict that specific RNA binding by this domain is diminished, potentially due to reduced RNA coordination in the predicted binding pocket. These changes in domain architecture between ZF1–2 and ZF3–4 are conserved in all three human MBNL homologs (MBNL1/MBNL2/MBNL3) (see Supplemental Figure S1C for sequence alignment). We predict that the differences in activity between these domains are maintained across MBNL1, MBNL2 and MBNL3 and potentially more broadly across MBNL proteins throughout metazoans (39).
Modular architecture of MBNL1 ZF domains provides a unique platform for RNA recognition
Although RBPs have a broad range of functions, they are often built from relatively few RNA binding domains. To increase the functionality and specificity of their target interactions, multiple RNA binding domains are frequently found in RBPs. A classic example of this is the Pumilio (PUF) family of RBPs, where up to 8 tandem domains that each recognize a single nucleotide can be combined in a single polypeptide chain to create a highly specific RNA interaction surface (55). In a similar manner we propose that the modular architecture of MBNL1 with its two tandem ZF pairs increases the protein–RNA binding surface. Our working model is that ZF1–2 drives splicing regulation through specific recognition of YGCY motifs and the ZF3–4 domain binds secondarily to a broader range of motifs to allow MBNL1 to recognize a wide range of substrates (Figure 8). Additionally, the domain organization and differences in RNA binding specificity between the ZF pairs may explain the relative levels of cooperativity observed in the dosing experiments (Figures 3–4, 7), assuming binding of the MBNL proteins correlates to splicing. One possible mechanism for cooperativity is that binding of one MBNL protein facilitates the binding of one or more additional MBNL proteins or other splicing factors to a pre-mRNA substrate. These additional binding events mediated by MBNL may shift splicing decisions over tighter protein gradients compared to MBNL-AA and MBNL-BB, which, in general, had less cooperative splicing curves.
This model of a modular RBP containing multiple domains, one for specific RNA recognition and the other with broader target binding, has been utilized by several other RBPs, including those containing CCCH zinc finger motifs. One example is the neuronal protein Unkempt, a highly conserved RBP that binds to its target mRNAs to reduce translation and is required for the establishment of neuronal morphology in development (56,57). Unkempt contains six CCCH ZF domains that form two tandem clusters, each with three ZFs (ZF1–3 and ZF4–6) (56,57). Both CLIP and structural data confirm that ZF1–3 binds to a UAG trinucleotide while ZF4–6 binds to a more variable U-rich motif (56,57). Mutational analysis of RNAs bound to Unkempt in vitro revealed that the UAG motif was mandatory for binding while alterations to the downstream U-rich element were more tolerated (57). These data suggest that in a manner similar to MBNL1 ZF1–2, Unkempt ZF1–3 drives RNA recognition via binding to the UAG sequence while binding to the less ‘specific’ U-rich motif by ZF4–6 allows for recognition of a wider array of RNA substrates in a manner similar to MBNL1 ZF3–4. The similar modes of MBNL1 and Unkempt RNA interactions indicate that this might be a common strategy for RNA binding proteins with ZF domains.
Engineered MBNL1s as protein therapeutics in neuromuscular disorders
The creation of designer RBPs has increased over the past several years as a means to modulate RNA function (58,59). Although the traditional methodology of engineering these proteins often focuses on combining domains to target a specific RNA sequence of interest, such as with the PUF proteins (58,59), we choose to utilize a different synthetic design strategy. We focused on enhancing the pre-existing activity and specificity of MBNL by re-combining its ZF domains. No such designer RBP has previously been created that focuses on enhancement of protein function via duplication of specific modular RNA binding domains. This design strategy may be the most effective for engineering RBPs as protein therapeutics in which the function of a target protein is decreased or absent such as in DM.
MBNL1 overexpression has been proposed as a therapeutic strategy in the DM field to ameliorate symptoms caused by the loss of free MBNL1 in CUG/CCUG RNA foci. Delivery of MBNL1 through adeno-associated virus (AAV) has been shown to rescue mis-splicing events in a DM1 mouse model and reverse disease associated symptoms in skeletal muscle, including myotonia (60). Additionally, MBNL1 overexpression has been shown to be well-tolerated in non-disease mice (61), suggesting that therapies designed to increase levels of free, active MBNL1 in the cell could be an effective strategy for treatment of DM. Delivery of a synthetic MBNL with increased activity, such as MBNL-AA, could be a powerful approach to correct disease-specific mis-splicing. The use of a synthetic MBNL as a protein therapeutic is potentially ideally suited for Fuchs Endothelial Corneal Dystrophy where the protein would only need to be delivered to single tissue, the eye (30). Our work to create a synthetic MBNL serves as a proof of principle that MBNL proteins tolerate domain reorganization and can be manipulated while retaining function. Further rational design strategies to modify MBNL could be utilized moving forward to continue to create a smaller, more stable, and higher activity synthetic MBNL for use in disease therapies. Overall, our studies indicate that engineered RNA binding proteins with improved splicing activity may represent a therapeutic avenue for DM and other microsatellite diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Jamie Purcell and Julia Oddo for their initial conception of this work as well as Jacob Gacke for his preliminary work on the plasmid dosing system. We would also like to thank members of the Berglund Lab and Center for NeuroGenetics for their helpful feedback and comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Florida (UF) [to J.A.B. and E.T.W.]; National Institutes of Health [GM121862 to J.A.B., OD017865 to E.T.W.]; Rosaria Haugland Graduate Research Fellowship [to M.A.H.]; Promise to Kate Graduate Research Fellowship [to M.A.H.]. Funding for open access charge: UF startup funds.
Conflict of interest statement. The University of Florida, J. Andrew Berglund and Melissa A. Hale have filed a provisional patent application for the use of synthetic MBNL proteins for treatment of repeat expansion diseases.
REFERENCES
- 1. Kalsotra A., Cooper T.A.. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011; 12:715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 4. Fu X.-D., Ares M.. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jangi M., Sharp P.A.. Building robust transcriptomes with master splicing factors. Cell. 2014; 159:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baralle F.E., Giudice J.. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell. Biol. 2017; 18:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pascual M., Vicente M., Monferrer L., Artero R.. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006; 74:65–80. [DOI] [PubMed] [Google Scholar]
- 8. Fernandez-Costa J.M., Llamusi M.B., Garcia-Lopez A., Artero R.. Alternative splicing regulation by Muscleblind proteins: from development to disease. Biol. Rev. 2011; 86:947–958. [DOI] [PubMed] [Google Scholar]
- 9. Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A.. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006; 15:2087–2097. [DOI] [PubMed] [Google Scholar]
- 10. Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A.. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dixon D.M., Choi J., El-Ghazali A., Park S.Y., Roos K.P., Jordan M.C., Fishbein M.C., Comai L., Reddy S.. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci. Rep. 2015; 5:9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang E.T., Ward A.J., Cherone J.M., Giudice J., Wang T.T., Treacy D.J., Lambert N.J., Freese P., Saxena T., Cooper T.A. et al. . Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015; 25:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blech-Hermoni Y., Ladd A.N.. RNA binding proteins in the regulation of heart development. Int. J. Biochem. Cell Biol. 2013; 45:2467–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang E.T., Cody N.A.L., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S. et al. . Transcriptome-wide regulationof pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012; 150:710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masuda A., Andersen H.S., Doktor T.K., Okamoto T., Ito M., Andresen B.S., Ohno K.. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci. Rep. 2012; 2:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osborne R.J., Lin X., Welle S., Sobczak K., O’Rourke J.R., Swanson M.S., Thornton C.A.. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009; 18:1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du H., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A. et al. . Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010; 17:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A. et al. . Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014; 56:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rau F., Freyermuth F., Fugier C., Villemin J.-P., Fischer M.-C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D. et al. . Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011; 18:840–845. [DOI] [PubMed] [Google Scholar]
- 20. Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T.. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992; 68:799–808. [DOI] [PubMed] [Google Scholar]
- 21. Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P.. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001; 293:864–867. [DOI] [PubMed] [Google Scholar]
- 22. Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D.. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002; 11:805–814. [DOI] [PubMed] [Google Scholar]
- 23. Fardaei M., Larkin K., Brook J.D., Hamshere M.G.. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001; 29:2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mankodi A., Urbinati C.R., Yuan Q.P., Moxley R.T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M.S., Thornton C.A.. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001; 10:2165–2170. [DOI] [PubMed] [Google Scholar]
- 25. Lee J.E., Cooper T.A.. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009; 37:1281–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klein A.F., Gasnier E., Furling D.. Gain of RNA function in pathological cases: focus on myotonic dystrophy. Biochimie. 2013; 93:2006–2012. [DOI] [PubMed] [Google Scholar]
- 27. Chau A., Kalsotra A.. Developmental insights into the pathology of and therapeutic strategies for DM1: Back to the basics. Dev. Dyn. 2015; 244:377–390. [DOI] [PubMed] [Google Scholar]
- 28. Meola G., Cardani R.. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim. Biophys. Acta. 2015; 1852:594–606. [DOI] [PubMed] [Google Scholar]
- 29. Daughters R.S., Tuttle D.L., Gao W., Ikeda Y., Moseley M.L., Ebner T.J., Swanson M.S., Ranum L.P.W.. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009; 5:e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du J., Aleff R.A., Soragni E., Kalari K., Nie J., Tang X., Davila J., Kocher J.-P., Patel S.V., Gottesfeld J.M. et al. . RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J. Biol. Chem. 2015; 290:5979–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Purcell J., Oddo J.C., Wang E.T., Berglund J.A.. Combinatorial mutagenesis of MBNL1 zinc fingers Elucidates distinct classes of regulatory events. Mol. Cell. Biol. 2012; 32:4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goers E.S., Purcell J., Voelker R.B., Gates D.P., Berglund J.A.. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010; 38:2467–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Teplova M., Patel D.J.. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008; 15:1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cass D., Hotchko R., Barber P., Jones K., Gates D.P., Berglund J.A.. The four Zn fingers of MBNL1 provide a flexibleplatform for recognition of its RNA bindingelements. BMC Mol. Biol. 2011; 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ho T.H., Charlet-B N., Poulos M.G., Singh G., Swanson M.S., Cooper T.A.. Muscleblind proteins regulate alternative splicing. EMBO J. 2004; 23:3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park S., Phukan P.D., Zeeb M., Martinez-Yamout M.A., Dyson H.J., Wright P.E.. Structural basis for interaction of the tandem zinc finger domains of human muscleblind with cognate RNA from human cardiac troponin T. Biochemistry. 2017; 56:4154–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Irion U. Drosophila muscleblind codes for proteins with one and two tandem zinc finger motifs. PLoS ONE. 2012; 7:e34248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oddo J.C., Saxena T., McConnell O.L., Berglund J.A., Wang E.T.. Conservation of context-dependent splicing activity in distant Muscleblind homologs. Nucleic Acids Res. 2016; 44:8352–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vicente-Crespo M., Pascual M., Fernandez-Costa J.M., Garcia-Lopez A., Monferrer L., Miranda M.E., Zhou L., Artero R.D.. Drosophila muscleblind is involved in troponin T alternative splicing and apoptosis. PLoS One. 2008; 3:e1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warf M.B., Berglund J.A.. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007; 13:2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S.. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004; 13:495–507. [DOI] [PubMed] [Google Scholar]
- 43. Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B. et al. . piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grammatikakis I., Goo Y.H., Echeverria G.V., Cooper T.A.. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011; 39:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tran H., Gourrier N., Lemercier-Neuillet C., Dhaenens C.M., Vautrin A., Fernandez-Gomez F.J., Arandel L., Carpentier C., Obriot H., Eddarkaoui S. et al. . Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J. Biol. Chem. 2011; 286:16435–16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edge C., Gooding C., Smith C.W.. Dissecting domains necessary for activation and repression of splicing by muscleblind-like protein 1. BMC Mol. Biol. 2013; 14:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kosaki A., Nelson J., Webster N.J.. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J. Biol. Chem. 1998; 273:10331–10337. [DOI] [PubMed] [Google Scholar]
- 48. Sen S., Talukdar I., Liu Y., Tam J., Reddy S., Webster N.J.G.. Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J. Biol. Chem. 2010; 285:25426–25437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hino S.-I., Kondo S., Sekiya H., Saito A., Kanemoto S., Murakami T., Chihara K., Aoki Y., Nakamori M., Takahashi M.P. et al. . Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007; 16:2834–2843. [DOI] [PubMed] [Google Scholar]
- 50. Gates D.P., Coonrod L.A., Berglund J.A.. Autoregulated splicing of muscleblind-like 1 (MBNL1) Pre-mRNA. J. Biol. Chem. 2011; 286:34224–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Philips A.V., Timchenko L.T., Cooper T.A.. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998; 280:737–741. [DOI] [PubMed] [Google Scholar]
- 52. Cleary J.D., Pearson C.E.. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003; 100:25–55. [DOI] [PubMed] [Google Scholar]
- 53. Wagner S.D., Struck A.J., Gupta R., Farnsworth D.R., Mahady A.E., Eichinger K., Thornton C.A., Wang E.T., Berglund J.A.. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 2016; 12:e1006316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fu Y., Ramisetty S.R., Hussain N., Baranger A.M.. MBNL1-RNA recognition: contributions of MBNL1 sequence and RNA conformation. ChemBioChem. 2011; 13:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell. Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murn J., Teplova M., Zarnack K., Shi Y., Patel D.J.. Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt. Nat. Struct. Mol. Biol. 2016; 23:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Murn J., Zarnack K., Yang Y.J., Durak O., Murphy E.A., Cheloufi S., Gonzalez D.M., Teplova M., Curk T., Zuber J. et al. . Control of a neuronal morphology program by an RNA-binding zinc finger protein, Unkempt. Genes Dev. 2015; 29:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mackay J.P., Font J., Segal D.J.. The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 2011; 18:256–261. [DOI] [PubMed] [Google Scholar]
- 59. Chen Y., Varani G.. Engineering RNA-binding proteins for biology. FEBS J. 2013; 280:3734–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kanadia R.N., Shin J., Yuan Y., Beattie S.G., Wheeler T.M., Thornton C.A., Swanson M.S.. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11748–11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chamberlain C.M., Ranum L.P.W.. Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum. Mol. Genet. 2012; 21:4645–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.