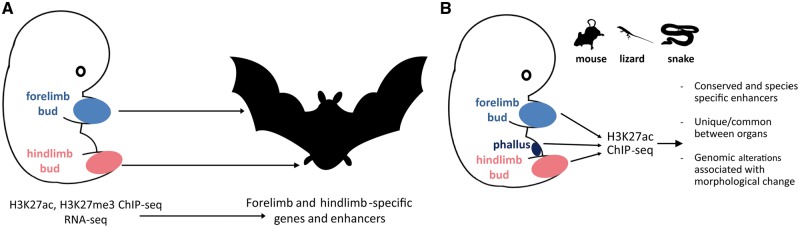
Figure 3.
Examples of using ChIP-seq to map GRNs in development and evolution. (A). Cataloging enhancers involved in patterning the morphology of bat limbs. The forelimb bud of bats develops into elongated webbed wings, while the hindlimb bud produces much shorter legs. H3K27ac and H3K27me3 ChIP-seq and RNA-seq were applied to developing limb buds to chart the developmental enhancer landscape involved in the specification of these structures [82]. (B) Understanding the role of limb-specific enhancers in snake evolution. Snakes lack legs, but H3K27ac ChIP-seq in mouse and lizard embryos and comparative genomics reveal that a substantial portion of limb enhancers are in fact conserved in snakes, one major reason for which is their role during phallus development, where a similar developmental program is deployed [83].