Figure 3.
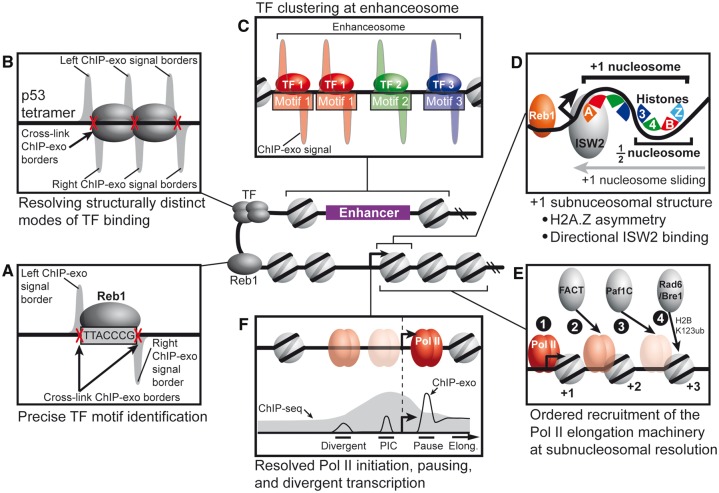
Models for how unique biological insights are revealed with the near base-pair resolution of ChIP-exo. (A) ChIP-exo signals delineate left and right protein–DNA cross-link borders, which enables precise TF motif identification. (B) ChIP-exo enables determination of structurally distinct modes of TF binding, such as the p53 tetrameric complex. (C) ChIP-exo distinguishes clustered TFs, such as the assembly of the enhanceosome. (D) Subnucleosomal structure of histone subunit–DNA contacts can be resolved by ChIP-exo. For example, the H2A.Z histone variant is asymmetrically incorporated into the +1 nucleosome. In addition, the ISW2 chromatin remodeler engages the +1 nucleosome on the promoter proximal side. (E) Ordered recruitment of Pol II elongation machinery at subnucleosomal resolution. (F) ChIP-exo resolves Pol II engaged at the PIC site, promotor proximal pause site and divergently oriented PIC.