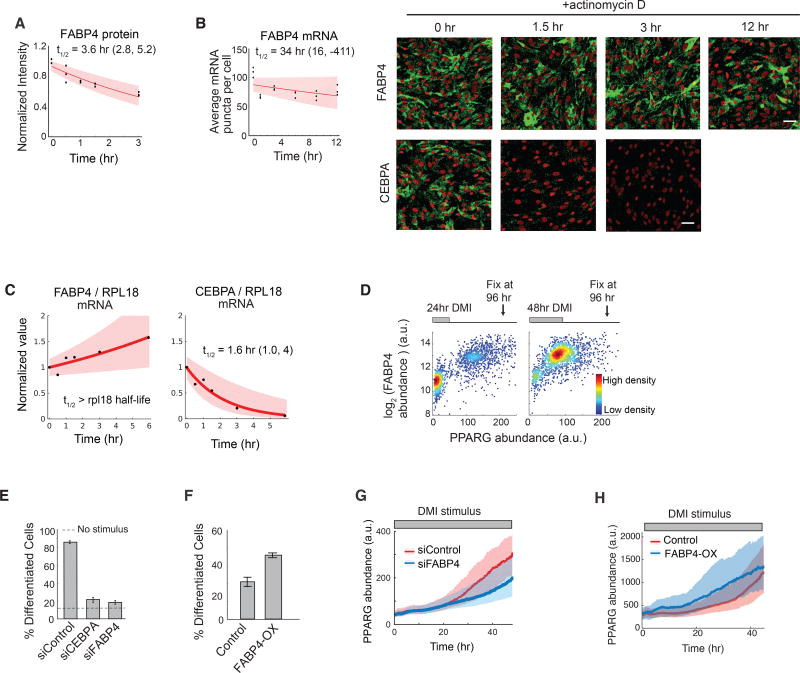
Figure 5. FABP4 is an example of a slow-degrading PPARG regulator that can mediate a slow increase in PPARG expression during adipogenesis.
(A) Half-life analysis of FABP4 protein as in Figure 2G.
(B) Half-life of FABP4 mRNA obtained using mRNA FISH as in Figure 2G. CEBPA mRNA FISH images are shown for comparison. Scale bars are 50 µm. See also Figure S6.
(C) Measurements using actinomycin D and RT-PCR show that the half-life of FABP4 mRNA is much longer than RPL18 mRNA, a known long-lived control. The half-life of CEBPA versus RPL18 mRNA is shown for comparison. First-order exponential decay curves were fitted to data. The shaded region represents the 95th confidence bounds. See also Figure S6.
(D) Single cell immunofluorescence analysis showing the mean nuclear PPARG and the mean cytosolic FABP4 values in each cell stimulated with DMI for 24 hours or 48 hours and fixed at 96 hours. Each scatter plot represents approximately 5000 cells and is colored based on cell density.
(E) Suppression of differentiation by siRNA-mediated depletion of FABP4 and CEBPA in OP9 cells.
(F) Overexpression of FABP4 increases differentiation even without DMI stimulation. OP9 cells were transfected with constructs expressing either mcherry(control) or mcherry-FABP4 and left in normal growth media for 5 days. No adipogenic stimulus was added.
(E–F) Percent of differentiated cells was quantitated as in Figure 1B. Barplots show mean +/− s.e.m. from 3 technical replicates, representative of 3 independent experiments.
(G–H) Live-cell imaging of citrine-PPARG cells transfected in (G) with FABP4-targeted or control siRNAs or in (H) with mcherry-FABP4 or control(mcherry). Cells were induced to differentiate using the standard DMI protocol. Plotted lines are population median traces with shaded regions representing 25th and 75th percentiles of approximately 700 cells per condition.