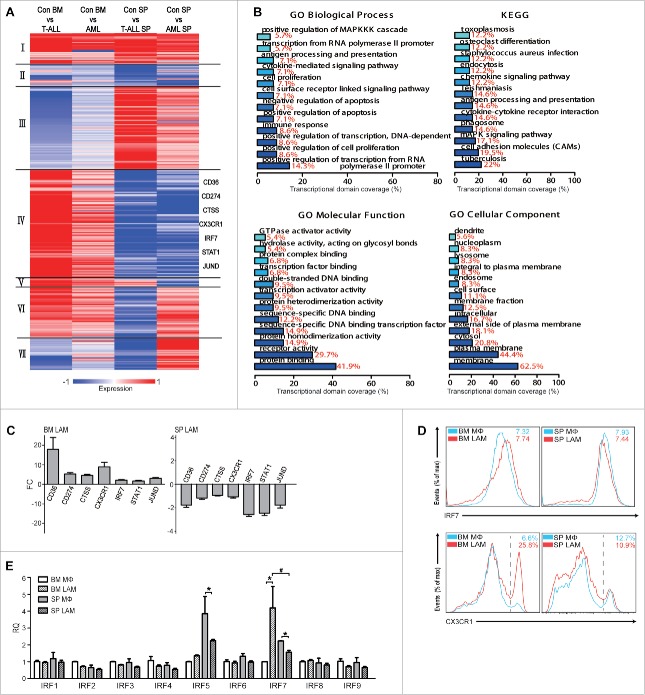
Figure 5.
Screening for key molecules governing differential evolution of LAMs in BM and SP. (A) K-mean clustering of 303 DEGs in BM and SP LAMs from leukemic microenvironment in T-ALL and AML mice. (B) GO annotation and KEGG pathway analysis were performed on DEGs, which were up-regulated in BM LAMs but down-regulated in SP LAMs. Each bar represents a significant annotation (P<0.05). The x axis shows the transcriptional domain coverage. (C) The genes up-regulated in BM LAMs but down-regulated in SP LAMs were validated by real time PCR. Fold change of those was analyzed against control macrophages from BM (left) or SP (right), respectively. (D) Expression of IRF7 and CX3CR1 in BM and SP LAMs from AML mice was detected by flow cytometry. The means of MFI ( × 104) or positive rates (in percentage) are indicated. (E) Expression of IRF family genes were detected by real-time PCR. The RQ value of control macrophages from BM was designated as 1.000. Data represent means (±s.d.). * P<0.05, vs. respective control; # P<0.05, SP LAMs vs. BM LAMs (Student's t-test). Data are representative of at least three independent experiments.