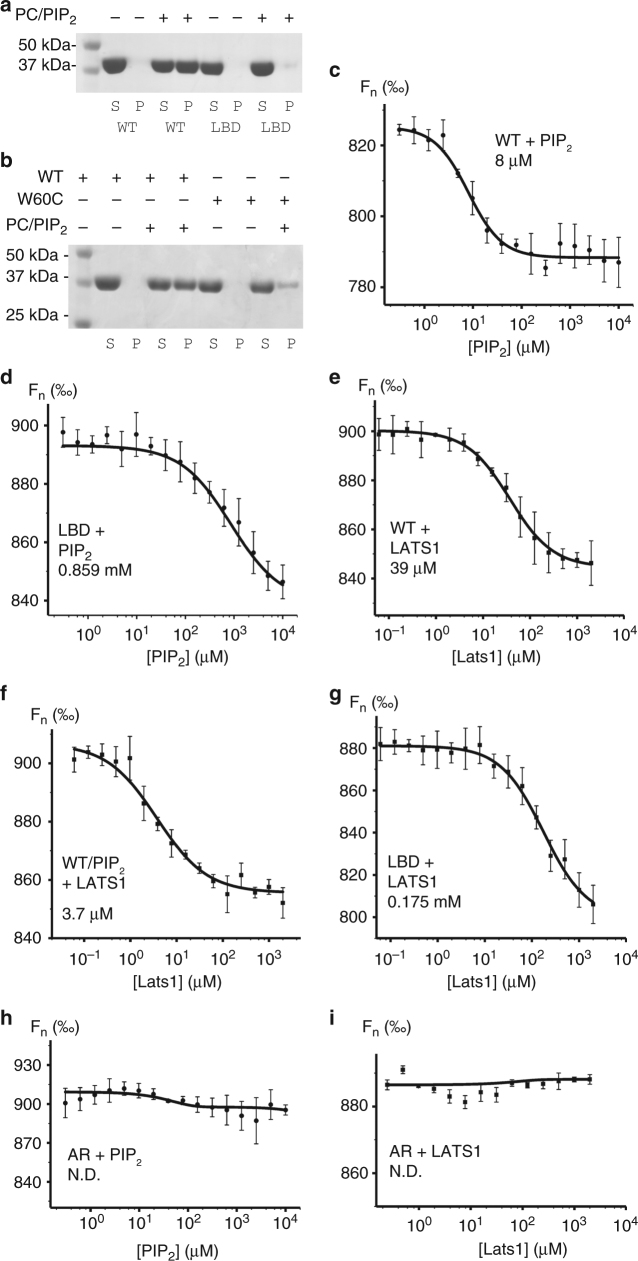
Fig. 3.
The conformation of neurofibromin 2 dictates its binding. a Lipid co-sedimentation analysis of the PIP2 binding to wild type and our lipid binding deficient mutant neurofibromin 2. wild-type neurofibromin 2 (residues 1–339) is soluble in the absence of PIP2 and pellets in the presence of PIP2. Mutant (T59V, W60E, R309Q, R310Q) neurofibromin 2 (residues 1–339) remains soluble in the absence and presence of PIP2. S supernatant, P pellet, WT wild type, LBD lipid binding deficient. b Lipid co-sedimentation analysis of the PIP2 binding to wild-type and disease-derived mutant neurofibromin 2. Wild-type neurofibromin 2 (residues 1–339) is soluble in the absence of PIP2 and pellets in the presence of PIP2. Mutant (W60C) neurofibromin 2 (residues 1–339) is soluble in the absence of PIP2, while a small fraction pellets in the presence of PIP2. S supernatant, P pellet, WT wild type. Microscale thermophoresis (MST) measurements show the binding of PIP2 to c wild-type full-length neurofibromin 2 (Kd = 8.02 ± 0.91 μM) or to d our lipid binding deficient (LBD) mutant (T59V, W60E, R309Q, R310Q; Kd = 859.23 ± 184.65 μM). MST measurements show binding of LATS1 (residues 69–100) to e wild type (Kd = 39.31 ± 4.25 μM), f the neurofibromin/PIP2 complex (Kd = 3.77 ± 0.72 μM), or g to our LBD neurofibromin 2 mutant (Kd = 175.54 ± 34.49 μM). No binding was observed for the artificially closed A585W-R588K (AR) mutants to h PIP2, or i LATS1. Error bars represent ±S.D., n = 3 (three independent measurements with the same laser power)