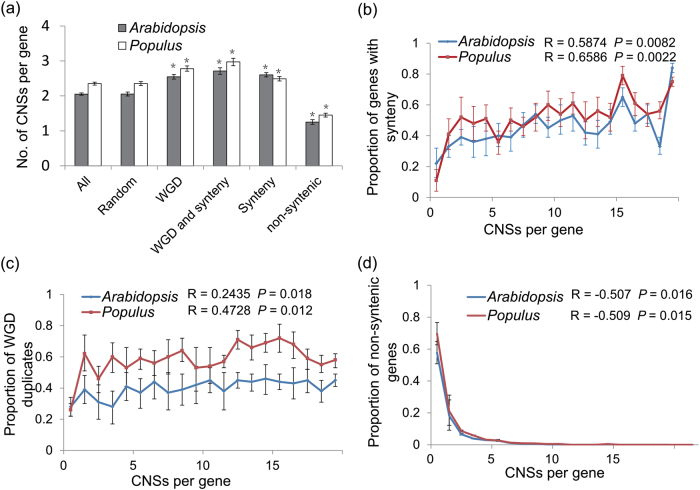
Fig. 4.
Retained paralogs and syntenic genes have more CNSs. a The mean number of CNSs linked to different syntenic, non-syntenic, and whole-genome duplication gene sets. In Populus: all (n = 41,335), synteny (n = 11,867), non-syntenic (n = 29,468), paralog retention (n = 18,164), paralog retention and synteny (n = 7804); and in Arabidopsis: all (n = 27,206), synteny (n = 9694), non-syntenic (n = 17,512), paralog retention (n = 8504), paralog retention and synteny (n = 4067) samples were used for the experiment. b The association between CNSs and synteny in Populus and Arabidopsis for CNSs. Each data point is calculated from at least 50 genes. c The association between CNSs and paralog retention in Populus and Arabidopsis for CNSs. d The association between CNSs and non-syntenic genes in Populus and Arabidopsis for CNSs. Each data point is calculated from at least 50 genes. Error bars, bootstrap-based 95% confidence intervals on the mean estimates. Both of two trends b, c represent a significant positive correlation between CNSs number and synteny/paralog retention, and d represent a significant negative correlation between CNSs number and non-syntenic genes conservation. where R is the Pearson correlation coefficient, and P is the upper tail probability that the true correlation is greater than zero