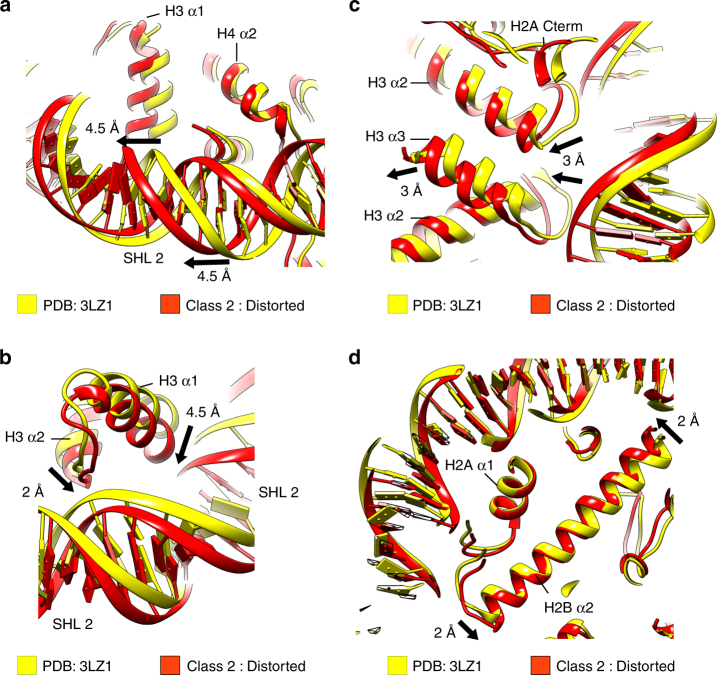
Fig. 2.
Structural rearrangements in the histone core. Comparison of the X-ray structure (PDB:3LZ1, yellow) and the Class 2 model (red). Arrows depict the direction of the helix movements. The degree of movement between X-ray structure and the Class 2 model is shown, rounded to half an Å. a Conformational rearrangement of H3 α1 and DNA at SHL 2. H3 α1 moves 4.5 Å in Class 2 when compared with the X-ray structure. DNA at SHL 1.5 moves 4.5 Å towards SHL 2. b Conformational rearrangement of the H3 α2, H3 α1, and DNA at SHL 2. The H3 α2 tilts by 2 Å at its N-terminal end. The H3 α1 moves 4.5 Å. H3 α1 and H3 α2 move toward each other and push the DNA at SHL 2.5 outward. c Conformational rearrangement of H3 α2, H3 α3, and DNA at the dyad. H3 α2 tilts and moves inward by 3 Å at its C-terminal end. H3 α3 also moves inward by 3 Å. Concomitantly, the DNA at the dyad moves more than 3 Å toward the centre of the nucleosome. d Conformational rearrangement of H2B α2. H2B α2 tilts by 2 Å toward the center of the nucleosome at SHL 3.5 and 2 Å away from the center at SHL 5.5. This pushes the DNA at SHL 5.5 outward and pulls the DNA at SHL 3.5 inward