Fig. 1.
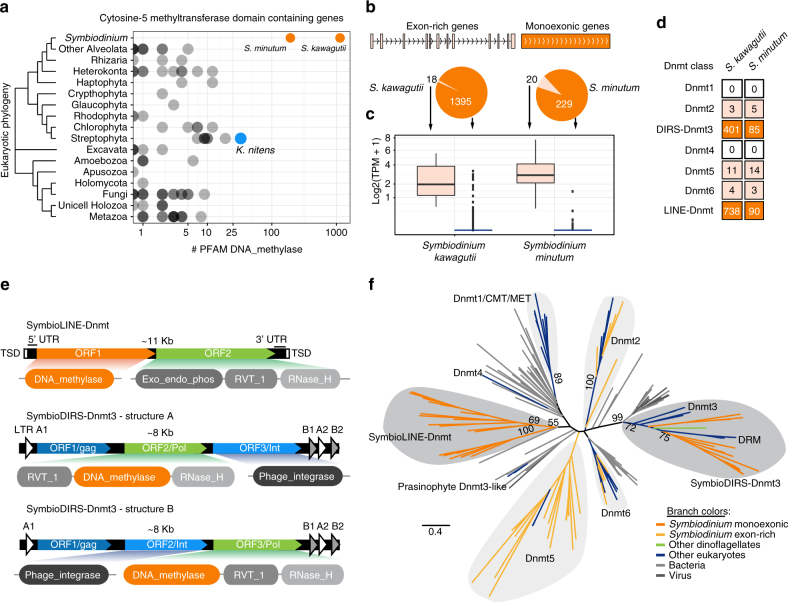
Symbiodinium genomes encode hundreds of DNMTs. a Number of DNMT genes encoded in eukaryotic lineages based on a data set of 96 genomes (Supplementary Data 1). Each dot corresponds to the number of genes containing a DNMT domain (PF00145, DNA_methylase) in a given genome. b Proportion of distinct DNMT gene architectures in Symbiodinium, pale shade for exon-rich, darker shade for monoexonic. c Transcriptional level of DNMT-encoding genes according to gene architecture in Symbiodinium. Boxplot centre lines are medians, box limits are quartiles 1 (Q1) and 3 (Q3), whiskers are 1.5 × interquartile range (IQR) and points are outliers. d Classification of DNMTs in Symbiodinium genomes, showing number of copies per family. Colour code reflects the type of gene architecture for each family: pale shade = exon-rich, darker shade = monoexonic. e Structure of Symbiodinium retrotransposons that encode for DNMTs and domain architectures of the open reading frames (ORFs). Domains named as in Pfam searches: Exo_endo_phos (PF03372) is an endonuclease domain, RVT_1(PF00078) is a reverse transcriptase, RNase_H (PF00075) is a RNase type H and Phage Integrase (PF00589) is a tyrosine recombinase. SymbioLINE-Dnmt show flanking tandem site duplications (TSD), conserved 5′ and 3′ UTR regions and lack Poly-A tails. SymbioDIRS-Dnmt3 show two sets of LTRs (represented by triangles A and B) in an arrangement typical of DIRS retrotransposons: A-coding region-B-A-B. f Maximum likelihood phylogenetic tree of a representative group of eukaryotic, bacterial and Symbiodinium DNMTs. Hundred parametric bootstrap replicates are shown as nodal supports for selected groups. Colour code indicates sequence taxonomic affiliation of each branch as shown in legend