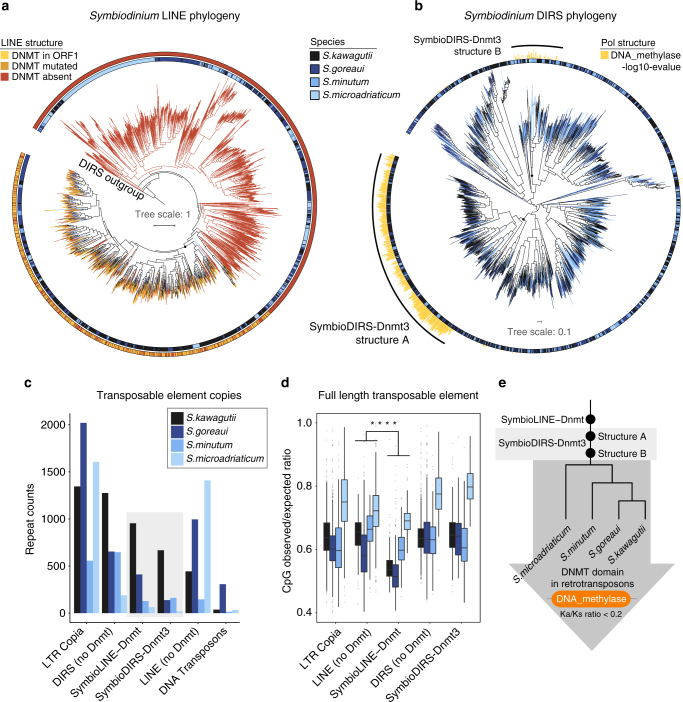
Fig. 2.
DNMT-retrotransposon evolutionary dynamics in Symbiodinium. a Maximum likelihood phylogenetic tree based on the ORF2 of Symbiodinium LINE retrotransposons, rooted using DIRS reverse transcriptases as an outgroup. The internal circle indicates species affiliation of each sequence and the outer circle indicates the architecture of the LINE element. DNMT mutated indicates that there are premature stop codons in ORF1 or ORF2. DNMT absent implies that there is no evidence of upstream ORF1 encoding a DNMT. b Maximum likelihood phylogenetic tree based on Pol ORF (retrotranscriptase and RNase_H domains) of DIRS-like retrotransposons. The inner circle indicates species affiliation and the barplot indicates the degree of DNMT conservation based on a hmmsearch alignment e-value. c Number of transposable element copies in each Symbiodinium species. d Comparison of the CG observed/expected ratio across all Symbiodinium retrotransposon types. SymbioLINE-DNMTs have fewer CGs than the rest (asterisks represent Wilcoxon one-sided rank-sum test p < 0.01) of retrotransposons in all four species, while SymbioDIRS-Dnmt do not show a significant difference when compared to DIRS elements without DNMT (p > 0.01). Boxplot centre lines are medians, box limits are quartiles 1 (Q1) and 3 (Q3), whiskers are 1.5 × interquartile range (IQR) and points are outliers. e Schematic species tree of Symbiodinium species showing the evolutionary origins of DNMT encoding retrotransposons in this lineage and the subsequent conservation of DNMT domain as shown in Supplementary Figs. 8 and 9. Phylogenetic relationships between Symbiodinium species is based on previous publications24, 38