Fig. 3.
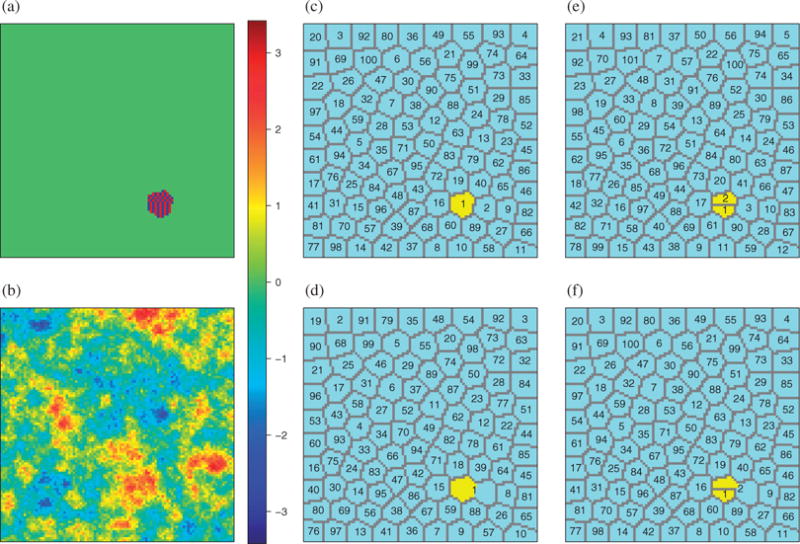
Model designs and configurations of different partitions on , the 100 × 100 equally spaced grid points in two-dimensional space [0, 1]2, in Setting 4 of the simulation study for spatial variable screening: (a) the true coefficient βj, which takes only three possible values, −3 in blue, 3 in red and 0 in green; (b) one set of simulated predictors Xj over space from a Gaussian random field on with covariance kernel exp(−10||s − s′||2) for s, s′ ∈ ; (c) the partition in Setting 4 to define the true nonzero coefficients, with βj ⧧ 0 if and only if , in yellow. Panels (c), (d), (e) and (f) respectively represent the optimal partition , the size-reduced partition , and two misspecified partitions, and , for the corresponding partition-based screening for selecting variables in Setting 4; in panels (c)–(f), is coloured yellow.
