Abstract
High-dimensional flow cytometry is proving to be valuable for the study of subtle changes in tumor-associated immune cells. As flow panels become more complex, detection of minor immune cell populations by traditional gating using biaxial plots, or identification of populations that display small changes in multiple markers, may be overlooked. Visualization of t-distributed stochastic neighbor embedding (viSNE) is an unsupervised analytical tool designed to aid the analysis of high-dimensional cytometry data. In this study we use viSNE to analyze the simultaneous binding of 15 fluorophore-conjugated Abs and one cell viability probe to immune cells isolated from syngeneic mouse MB49 bladder tumors, spleens, and tumor-draining lymph nodes to identify patterns of anti-tumor immune responses. viSNE maps identified populations in multidimensional space of known immune cells, including T cells, B cells, eosinophils, neutrophils, dendritic cells, and NK cells. Based on the expression of CD86 and programmed cell death protein 1, CD8+ T cells were divided into distinct populations. Additionally, both CD8+ T cells and CD8+ dendritic cells were identified in the tumor microenvironment. Apparent differences between splenic and tumor polymorphonuclear cells/granulocytic myeloid-derived suppressor cells are due to the loss of CD44 upon enzymatic digestion of tumors. In conclusion, viSNE is a valuable tool for high-dimensional analysis of immune cells in tumor-bearing mice, which eliminates gating biases and identifies immune cell subsets that may be missed by traditional gating.
The altered expression of many markers on immune cells in mouse tumor models has created a need for new methods to analyze high-dimensional data. New computational techniques that use dimension reduction to aid in the analysis of high-dimensional cytometry data have been developed recently (reviewed in Refs. 1–3). One such dimension-reduction algorithm is referred to as visualization of t-distributed stochastic neighbor embedding (viSNE) (4) or t-distributed stochastic neighbor embedding (tSNE) (5). viSNE data can be visualized by overlaying heat maps or density plots to generate a third dimension of data representation (4). Each point on the viSNE map represents an individual cell that has been separated into spatially resolved populations based on the totality of markers that they express. However, proximity in two dimensions between distinct populations is not an indicator of similarity, because two-dimensional viSNE maps are derived from high-dimensional data (4). Although mainly used as a tool for the analysis of complex mass cytometry data (6–10), tSNE or viSNE (11, 12) can be useful for interpreting high-dimensional traditional flow cytometry data. In this study we use viSNE to analyze the simultaneous binding of 16 fluorophores (15 surface markers and a live/dead indicator) to immune cells isolated from murine bladder cancers or melanomas as well as tumor-draining lymph nodes (dLN) and spleens.
Bladder cancer impacts predominantly older men and is the fifth most common cancer in the United States, representing almost 5% of all new cancer cases each year (13). The MB49 cell line is a useful in vivo murine model of bladder cancer that has several similarities to human disease (14). MB49 tumors in mice are sensitive to treatment with the antituberculosis vaccine bacillus Calmette–Guérin (15, 16), an immunotherapy for treatment of early-stage bladder cancer that is thought to activate the immune system to slow tumor growth. To evaluate different aspects of disease, MB49 cells can be administered to mice via various routes, including transurethral, s.c. or i.v., leading to development of tumors and spontaneous lung metastasis within 3 wk (14, 16–18).
In the current study, C57BL/6 mice were injected s.c. with syngeneic MB49 cells. Following tumor growth for 21 d, immune cells in solid tumors, spleens, and tumor dLN were evaluated by flow cytometry. The generation of two-dimensional viSNE plots with heat maps aided the identification of populations in known immune cell subsets. viSNE maps separated immune cells into distinct populations that consist of the expected primary immune cell populations and varied in frequency among tissues. viSNE readily identified immune cell subsets that were frequent (i.e., granulocytes) or infrequent in tumors (i.e., B cells). Applying viSNE to subsets of immune cells did not impact the ability of the algorithm to identify rare populations, including CD8+ dendritic cells (DCs).
Materials and Methods
Mice
Female wild type (C57BL/6J) mice were obtained from the Jackson Laboratory. Animals were used at 6–8 wk of age for individual experiments. All experiments involving mice were performed according to the La Jolla Institute guidelines for laboratory animals and were approved by the Institutional Animal Care and Use Committee.
Cell lines and tumor growth
Dr. D. Theodorescu of the University of Colorado kindly provided us with the mouse urothelial carcinoma cell line MB49 originally produced by Dr. T. Ratliff of Purdue University. MB49 cells were confirmed to be of mouse origin and tested negative for evidence of cross-species contamination and pathogen contamination by IDEXX BioResearch (Columbia, MO). B16F10 melanoma cells obtained from the American Type Culture Collection were confirmed to be pure by IDEXX BioResearch. MB49 and B16F10 cells were cultured in R5F (RPMI 1640 medium containing 10% heat-inactivated FBS, 50 U/ml penicillin, 50 μg/ml streptomycin). Upon reaching 60–80% confluence, 100,000 cells were injected s.c. into the right flank. Tumor volumes were measured using digital calipers, and calculated as height × width2/2, at least two times a week. On day 21 post inoculation, mice were euthanized, and tumors and organs were harvested using an aseptic technique. The average MB49 tumor volume was 212 mm3 and the average B16F10 tumor volume was 391 mm3.
Flow cytometry
Spleens and right inguinal tumor dLN were disaggregated without enzyme exposure by being pressed through a 70 μm filter. RBCs were lysed in RBC Lysis Buffer according to the manufacturer’s protocol (Bio-Legend). Tumor samples were homogenized using the Miltenyi mouse tumor dissociation kit according to the manufacturer’s guidelines. In some cases, spleen cells were incubated with tumor dissociation buffer to evaluate the effects of proteolytic enzymes on marker expression. Cells were resuspended in PBS + 0.5% FBS and counted using a Countess cell counter (Thermo Fisher Scientific). Normalized numbers of cells were stained for 30 min at 4°C using LIVE/DEAD blue fixable dead cell stain (Thermo Fisher Scientific), and a 1:100 dilution of a full panel (Table I) or a subset (Table II) of Abs. Samples were acquired on a BD LSR Fortessa flow cytometer.
Table I.
Ab clones and fluorophores
| Ab | Clone | Fluorophore | Source |
|---|---|---|---|
| CD16/32 | 93 | eBioscience | |
| CD45 | 30-F11 | Alexa Fluor 488 | BioLegend |
| TCRβ | H57-597 | PerCP/Cy5.5 | BioLegend |
| CD4 | RM4-5 | BV 510 | BioLegend |
| CD8 | 53-6.7 | APC-eFluor 780 | eBioscience |
| CD44 | IM7 | Alexa Fluor 700 | BioLegend |
| CD25 | PC61 | PE Dazzle | BioLegend |
| CD64 | X54-5/7.1 | Alexa Fluor 647 | BioLegend |
| CD11b | M1/70 | BV 421 | BioLegend |
| CD11c | N418 | BV 570 | BioLegend |
| Ly6C | HK1.4 | PE-Cy7 | BioLegend |
| Ly6G | 1A8 | BV 711 | BioLegend |
| NK1.1 | PK136 | PE | BioLegend |
| MHC II | M5/114.15.2 | BV 785 | BioLegend |
| PD-1 | 29F.1A12 | BV 605 | BioLegend |
| CD86 | GL-1 | BV 650 | BioLegend |
Abs were diluted 1:100 and incubated with cells from tumors, dLN, and spleen.
Table II.
Ab clones and fluorophores used for PMN and gMDSC
| Ab | Clone | Fluorophore | Source |
|---|---|---|---|
| CD16/32 | 93 | eBioscience | |
| CD45 | 30-F11 | PerCP | BD Bioscience |
| CD44 | IM7 | Alexa Fluor 700 | BioLegend |
| CD11b | M1/70 | BV 421 | BioLegend |
| Ly6C | HK1.4 | APC | BioLegend |
| Ly6G | 1A8 | PE-Cy7 | BioLegend |
Abs used to detect CD11b and CD44 staining of Ly6C+Ly6G+ cells.
Analysis of flow cytometry data
Flow cytometry data were analyzed using Cytobank software (Cytobank, Santa Clara, CA). viSNE was run using default Cytobank parameters (iterations = 1000, perplexity = 30, θ = 0.5). A range of 600 to >100,000 live cells was acquired. Samples were randomly downsampled to 500–6000 events per sample and analysis was run on equal numbers of events per sample. The range in events was determined by the sample with the fewest events acquired. In each figure, all samples were derived from the same viSNE run. For consistency, corresponding supplemental figures analyzed a similar number of events per sample. Individual flow cytometry standard files from each viSNE run were combined into a single flow cytometry standard file to assist in defining spatially distinct populations using the concatenation tool. viSNE heat maps show fluorescent intensity of each marker for each event. Scales on the heat maps are individually generated for each surface marker from low to high expression. Two samples from the dLN of mice with MB49 tumors were excluded from analysis due to low viability and cell counts. All dLN samples were excluded from Fig. 3, Supplemental Fig. 3, and Supplemental Fig. 4 due to low numbers of Ly6C+ Ly6G+ cells acquired from this site. Fig. 2 heat maps show the average median fluorescent intensity (MFI) for each population isolated from each tissue and were generated using the conditional formatting tool in Microsoft Excel.
FIGURE 3.
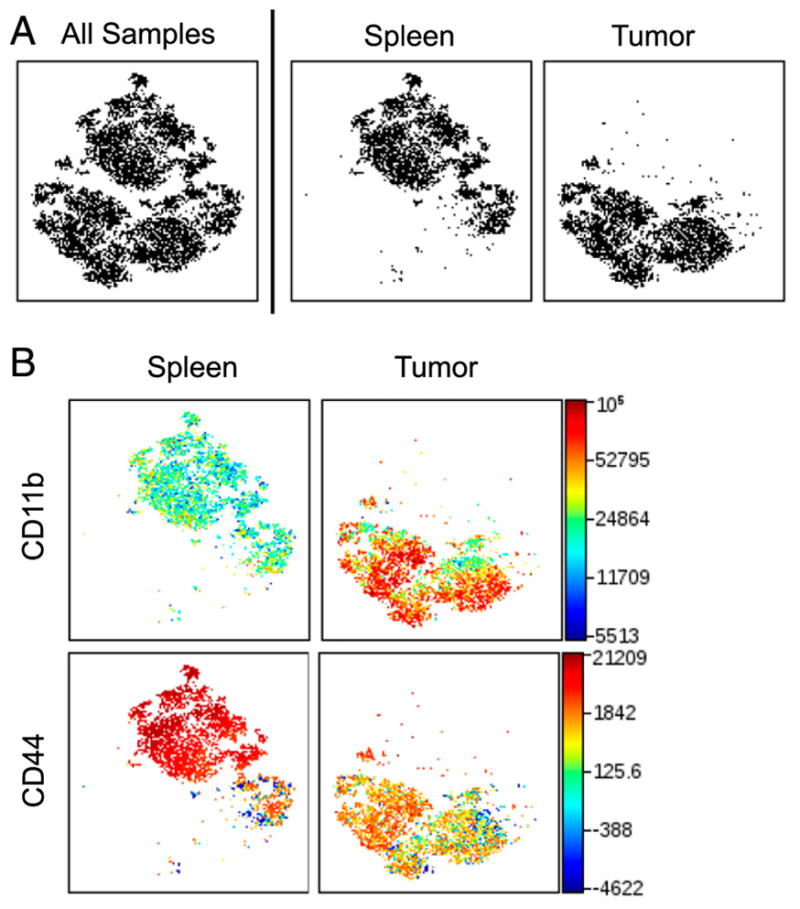
CD11b and CD44 expression define distinct populations of Ly6C+Ly6G+ cells. viSNE analysis of murine immune cells from MB49 tumors and spleens run on 500 live CD45+Ly6C+Ly6G+ single cells from each sample. (A) viSNE map showing concatenated flow cytometry standard files for all samples, spleen, and tumor. (B) Heat maps of CD44 and CD11b for all samples overlaid on the viSNE map, n = 9. All samples = 9000 events, spleen and tumor = 4500 events.
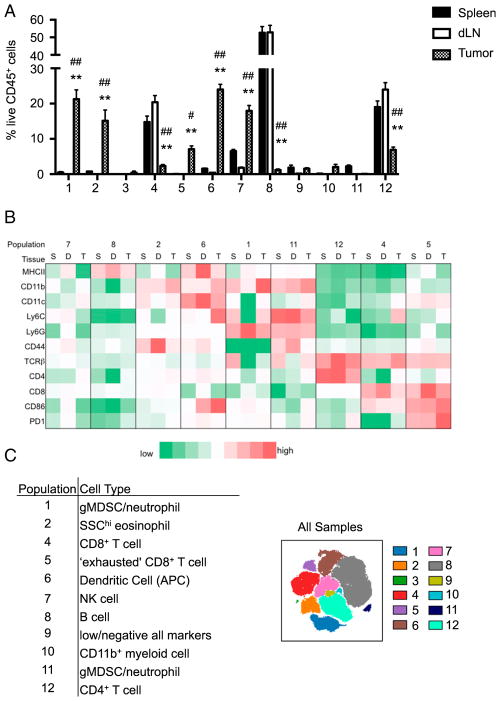
FIGURE 2.
Identification of viSNE-defined populations. viSNE analysis of murine immune cells in spleens, dLN, and MB49 tumors run on 6000 live CD45+ single cells from each sample. (A) Relative proportions of immune cell populations defined in Fig. 1B isolated from the tumor, spleen, and dLN. **p < 0.0001 when comparing spleen and tumor, #p = 0.001, ##p < 0.0001 when comparing dLN and tumor. The p values were derived by two-way ANOVA. (B) MFI of surface markers for each immune cell population isolated from the spleen (S), dLN (D), and tumor (T). (C) Predicted cell types based on surface marker expression and the frequency of cell populations among tissues.
Statistics
The p values were derived by two-way ANOVA with multiple comparisons and Tukey post hoc tests or paired Student t test using GraphPad Prism v6.0 software. Differences are considered statistically different when p ≤ 0.01.
Results
viSNE identifies distinct immune cell populations
Mice were euthanized 21 days after s.c. inoculation with 105 syngeneic MB49 bladder cancer cells, and cells derived from tumors, spleens, and dLN were isolated. Single-cell suspensions from each organ were stained with a mixture of 6 or 16 fluorophores (Tables I, II) and samples were acquired on a BD LSR Fortessa flow cytometer. Using Cytobank software, cells were gated on live CD45+ singlets then the viSNE algorithm analyzed 6000 events per sample. viSNE plots are shown in two dimensions with axes identified by tSNE-1 and tSNE-2 and each dot representing a single cell positioned in the multidimensional space (Fig. 1A). Individual flow cytometry standard files were concatenated into single flow cytometry standard files (Supplemental Fig. 1) from which 12 spatially distinct populations were identified (Fig. 1B). Similar spatially distinct immune cell populations were generated in response to B16F10 melanoma (Supplemental Fig. 2).
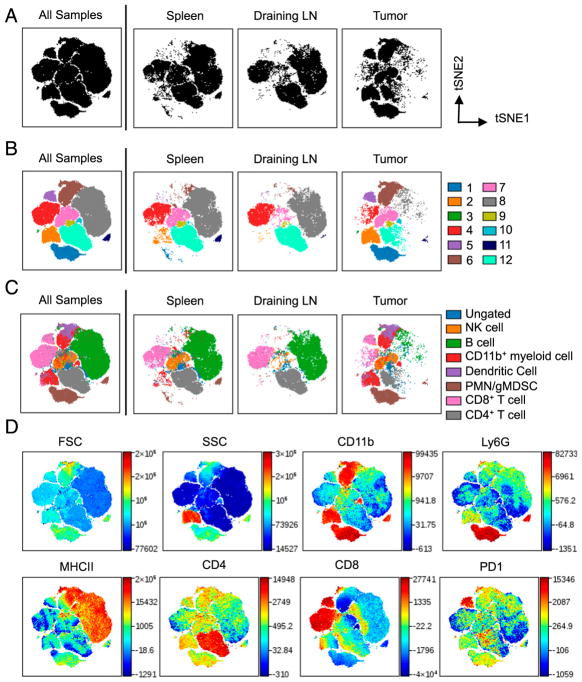
FIGURE 1.
viSNE defines populations of distinct immune cells. viSNE was used for the analysis of murine immune cells in spleen, dLN, and MB49 tumors. Cells were stained with 16 markers and measured by flow cytometry. viSNE analysis was run on 6000 live CD45+ single cells per sample using all surface markers. (A) viSNE map showing concatenated flow cytometry standard files for all samples, and individual spleen, dLN, and tumor sample files. (B) viSNE maps define 12 spatially distinct cell populations. (C) Overlay of manually gated cell populations on to viSNE plots. (D) Forward light scatter, SSC, and fluorescent intensity of CD11b, Ly6G, MHC II, CD4, CD8, and PD-1 for all samples overlaid on viSNE map. Spleen and tumor, n = 9. dLN, n = 7. All samples = 150,000 events; spleen and tumor = 54,000 events; dLN = 42,000 events.
To help identify cell populations, traditional biaxial gating strategies based on 1–3 surface markers were used: CD4+ T cells (TCRβ+CD4+), CD8+ T cells (TCRβ+CD8+), CD11b+ myeloid cells (CD11b+), DCs (CD11b+CD11c+), NK cells (NK1.1+), and polymorphonuclear cells/granulocytic myeloid-derived suppressor cells (PMN/gMDSC) (CD11b+Ly6C+Ly6G+). Overlaying immune cell populations identified by traditional gating strategies on to viSNE plots (Fig. 1C) and comparing them to viSNE heat maps (Fig. 1D) showed similarities between heat map intensities and surface markers used to define immune cell populations. Overlaid populations on viSNE maps show expected populations in a tissue-dependent manner. The spleens and dLN were composed primarily of B cells and T cells, with the spleen also containing granulocytes, CD11b+ myeloid cells, and NK cells (Fig. 1C) (19). In contrast to the spleen and lymph nodes, immune cells in the tumor microenvironment consisted primarily of CD11b+ myeloid cells with smaller proportions of T and B cells (Fig. 1C).
Biaxial gating failed to identify some CD45+ cells in viSNE plots, leaving a population that is ungated (shown in blue in Fig. 1C). The presence of unidentified cells confirms previous reports that traditional gating strategies do not account for all immune cells (4). Additionally, some traditionally gated immune cells, including CD11b+ cells (red), CD8+ T cells (pink), and PMN/gMDSC (brown), occupy multiple spatially distinct populations in the viSNE plot (Fig. 1C). To account for unidentified cells and immune cells that are found in multiple populations, further analysis was undertaken on viSNE-defined populations identified in Fig. 1B.
Using MFI and tissue distribution to identify viSNE-defined populations
Gating on each viSNE-defined population, relative proportions (Fig. 2A) and MFI (Fig. 2B) of each population isolated from the spleen, dLN, and tumors were calculated. MHC class II (MHC II) is expressed on APCs, including B cells, macrophages, and DCs (20), that present Ag peptides to CD4+ T cells. The primary cells that express MHC II are populations 6 and 8, with the highest expression on cells isolated from the dLN (Fig. 2B). Due to their expression of MHC II, populations 6 and 8 most likely consist of APCs. Similar to MHC II, CD86 is highly expressed by APCs and provides costimulation required for naive T cell activation. Population 6 expresses CD86, predominantly in the dLN and tumors, whereas population 8 does not express CD86 (Fig. 2B). Because of the high proportion of population 8 in the spleen and dLN (Fig. 2A) and their high expression of MHC II but not CD86 (Fig. 2B), this population consists of B cells (Fig. 2C). Based on surface marker expression, viSNE readily distinguished B cells despite the absence of a selective marker. In addition to the expression of MHC II and CD86, population 6 expresses high levels of CD11b and CD11c (Fig. 2B), which identifies this population as DCs (Fig. 2C) (21, 22). However, not all cells in population 6 express CD11c or are defined as DCs using biaxial gating strategies (Fig. 1C); therefore, some cells in this population may be macrophages that are not resolved into a distinct population. To suppress functional T cell responses, some DCs respond by expressing programmed cell death protein 1 (PD-1) (23, 24). As expected, PD-1 was observed on the surface of DCs isolated from the tumor dLN (Fig. 2B), although expression was not as high as in population 5. Significant expression of MHC II, CD86 and PD-1 by DCs in the dLN is consistent with high levels of Ag presentation to T cells at this site.
PD-1 is an inhibitory receptor that is upregulated on T cells activated through their TCR and costimulatory molecules, and functions to reduce T cell proliferation and inflammatory cytokine production. Based on expression of TCRβ, CD4, and CD8, population 12 defines CD4+ T cells and populations 4 and 5 define CD8+ T cells (Fig. 2B, 2C). In cancer, Ag-specific CD8+ T cells have been shown to express high levels of PD-1, which results in functionally impaired or “exhausted” CD8+ T cells (25). Additionally, expression of CD86 on T cells can be induced in response to IL-2 (26) produced during inflammation. Due to significant expression of PD-1 and CD86 on population 5 (Fig. 2B), these cells are classified as exhausted CD8+ T cells (Fig. 2C). As expected, exhausted CD8+ T cells are predominantly found in tumors (Fig. 2A). In contrast to population 5, population 4 does not express PD-1 or high levels of CD86 (Fig. 2B), enabling the generation of a spatially distinct population of nonexhausted CD8+ T cells. Compared to CD8+ T cells, CD4+ T cells were not found to express CD86 (Fig. 2B), but tumor-associated CD4+ T cells do express some PD-1 (Fig. 2B).
Differentiating between granulocytes
Several distinct populations of side scatter (SSC) high granulocytes were identified in Fig. 1, including populations 1, 2, and 11. Examples of granulocytes include neutrophils, eosinophils, basophils, and gMDSC. Population 2 exhibited a greater SSC profile compared with populations 1 and 11 (Fig. 1D), which readily identifies this population as eosinophils. Population 2 also expresses CD11b and CD44 (Fig. 2B). SSChiCD11b+ eosinophils were also found in B16F10 melanomas (Supplemental Fig. 2C), as previously described (27). Tumor eosinophils are known to promote recruitment of tumor-specific CD8+ T cells through the production of chemokines (28). However, the influence of eosinophils on clinical prognosis is tumor specific, with improved prognosis in bladder cancer and poor prognosis in melanoma (reviewed in Ref. 29).
Compared to eosinophils, populations 1 and 11 have a lower SSC profile and express CD11b, Ly6G, and Ly6C (Fig. 2B). These surface markers identify both PMN and gMDSC (30–36). In some reports, PMN and gMDSC cannot be resolved and are referred to as PMN-MDSC. The greatest difference between populations 1 and 11 in mice harboring MB49 tumors is the expression of CD44, which is high on population 11 but low on population 1 (Fig. 2B). Similarly, Ly6C+Ly6G+ cells isolated from melanomas expressed less CD44 than cells derived from the spleen (Supplemental Fig. 3). CD44 is a cell-surface glycoprotein involved in cell to cell interactions such as cell adhesion and migration. CD44 has been shown to be highly expressed on the surface of PMN (37) and is shed upon cell activation (38). To further characterize PMN/gMDSC populations, a separate viSNE analysis was run on sorted spleen and tumor CD45+Ly6C+Ly6G+ cells. As shown in Fig. 2, the main difference between these populations is their expression of CD44. CD44hi cells are primarily found in the spleen, whereas the CD44lo cells are predominantly found in tumors (Fig. 3). Variations in CD44 expression between the spleen and tumor is independent of whether Ly6C+Ly6G+ cells were isolated from the spleens of healthy or tumor-bearing mice (Supplemental Fig. 4). Of note, most Ly6C+Ly6G+ cells isolated from tumors express more CD11b than cells isolated from the spleen (Fig. 3B). CD11b (Mac-1) is an integrin whose expression on myeloid cells, including DCs (39) and PMN (40), is upregulated in response to cell simulation. Differential expression of CD11b suggests these populations may undergo location-dependent activation. However, no phenotypic difference was observed between CD44+ and CD44lo Ly6C+Ly6G+ cells isolated from the spleen or tumors (data not shown).
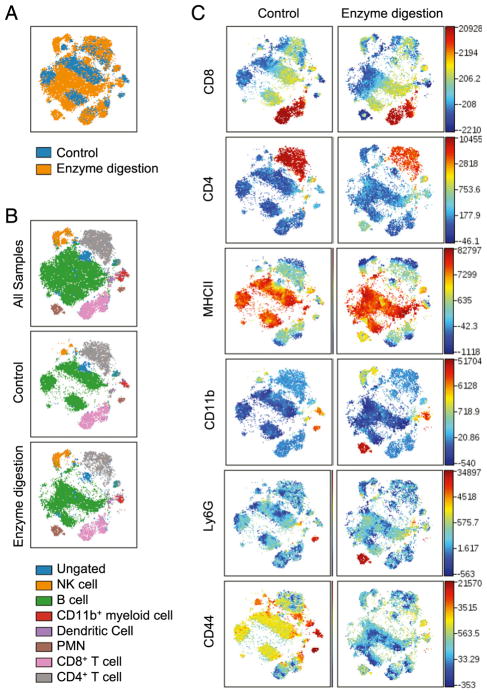
One significant difference between Ly6C+Ly6G+ cells purified from the spleen and tumors is the procedure used for their isolation. Isolation of tumor-infiltrating leukocytes requires both mechanical and enzymatic digestion, whereas spleen cells are mechanically disaggregated. To determine the effect of enzymes on immune cell marker expression, naive splenocytes were treated with the same enzyme mixture used to digest tumors. Splenocytes treated with digestion enzymes showed similar expression of most markers and little impact was observed on the localization of most viSNE-defined populations (Fig. 4). The notable exception is CD44, whose expression is significantly reduced by enzymatic digestion (Fig. 4C). Differences in CD44 expression by PMN (brown), NK cells (orange), and B cells (green) led to the identification of distinct populations in the viSNE map (Fig. 4B). Similar to what was observed in Fig. 1C, PMN show the greatest separation in high-dimensional space after enzymatic digestion (Fig. 4B). These findings lead us to conclude that the apparent differences between splenic and tumor PMN/gMDSCs are likely due to the loss of CD44 upon the enzymatic digestion of tumors.
FIGURE 4.
CD44 expression is sensitive to enzymatic digestion. viSNE analysis of naive splenocytes prepared either by mechanical or enzymatic disaggregation. Analysis was run on 6000 live CD45+ single cells per sample using all surface markers. (A) Overlaid viSNE maps showing concatenated flow cytometry standard files for all mechanical (control) or enzymatically digested splenocytes. (B) Overlay of manually gated cell populations on to concatenated viSNE plots. (C) Heat maps of markers in all samples overlaid on viSNE maps, n = 3. Splenocytes = 18,000 events; all = 36,000 events.
viSNE identifies rare cell populations
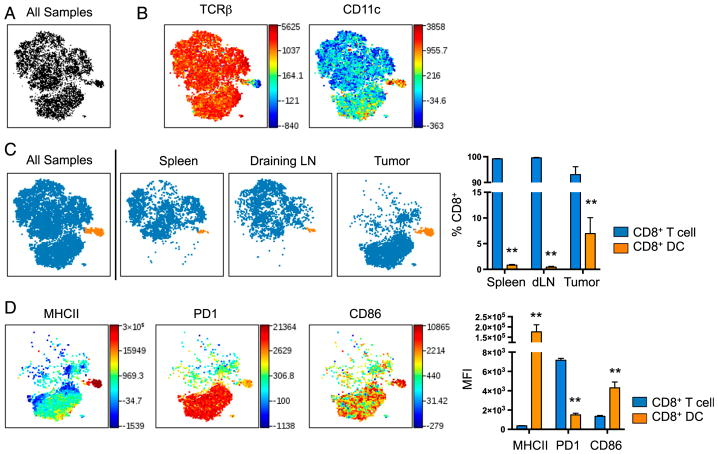
One benefit of viSNE is that it has been validated to identify rare populations representing as little as 0.25% of total events (4). In Fig. 1D, a small subset of population 6 can be seen to express CD8. Because population 6 was identified as DCs (Fig. 2), this subset of DCs may consist of CD11c+CD8+ DCs that cross-present Ag to CD8+ T cells via MHC class I more effectively than CD8− DCs (41) and endocytose dying cells (42). To focus the viSNE analysis on cells only expressing CD8, a separate analysis was run (Fig. 5A). As expected, a majority of CD8+ cells were TCRβ+ (Fig. 5B) and a majority of tumor-associated CD8+ T cells reproducibly expressed high levels of PD-1 (Fig. 5D). One population in the viSNE plot was TCRβ− CD11c+ (Fig. 5B). CD11c+CD8+ DCs can be found in the spleen, dLN, and tumors, with the highest relative frequency in tumors (Fig. 5C). Further characterization of CD8+ DCs revealed elevated expression of MHC II and CD86 relative to CD8+ T cells (Fig. 5D) (39).
FIGURE 5.
viSNE identifies a subset of tumor CD8+ cells that are CD11c+. viSNE analysis of murine immune cells in MB49 tumors, spleens, and dLN was run on 500 live CD45+CD8+ single cells from each sample. (A) viSNE map showing concatenated flow cytometry standard files for all samples. (B) Heat maps of markers in all samples overlaid on viSNE maps. (C) viSNE maps separated into two spatially distinct populations based on their expression of TCRβ and CD11c. A bar graph shows the percentages of each CD8 population among tissues. (D) Heat maps of markers in all samples overlaid on viSNE maps and quantified in bar graphs. Spleen and tumor, n = 9; dLN, n = 7. All samples = 12,500 events; spleen and tumor = 4500 events; dLN = 3500 events. **p < 0.001 by paired Student t test.
Discussion
In the current study, we describe the use of viSNE with fluorescence-based flow cytometry for high-dimensional analysis of immune cell populations in primary tumors and peripheral tissues of tumor-bearing mice. High-dimensional analysis of live cells combined with traditional analysis of biaxial-gated immune cell populations revealed the presence of the major expected immune cell populations. Gating on cell populations identified by viSNE allowed for resolution of subpopulations that differ in their expression of multiple activation markers. Additionally, applying viSNE analysis to subsets of immune cells did not impede its ability to define rare populations or to distinguish between populations that share similar surface markers.
viSNE identified variable frequencies of immune cell populations in a tissue-dependent manner, similar to what has previously been reported using traditional biaxial gating strategies. The spleen and dLN were primarily composed of adaptive immune cells (T cells and B cells) with few innate immune cells (macrophages, DCs, NK cells, and PMN) whereas leukocytes found in the tumor microenvironment consisted primarily of innate immune cells. viSNE revealed expected differences in the expression of MHC II, CD86, and PD-1 by immune cells. Ag presenting cells such as B cells, macrophages, and DCs are the primary cells that express MHC II and CD86. CD8 T cells isolated from tumors expressed high levels of PD-1 and accounted for a spatially distinct population in the viSNE map. Therefore, viSNE identified expected immune cell populations and readily revealed tissue-dependent differences in activation and exhaustion marker expression.
A unique feature of viSNE is its ability to reproducibly identify immune cell populations in the absence of selective markers (4). Even in the absence of a B cell-specific marker in the flow panel, viSNE was able to identify B cells in the spleen and dLN that were TCRβ−CD11b−MHCII+. This combination of cell surface markers distinguished B cells from other APCs such as macrophages and DCs (20). Additionally, viSNE identified a population of eosinophils that were SSChiCD11b+Ly6C−Ly6G−MHCII−. This combination of cell surface markers readily distinguished eosinophils from other granulocytes, such as PMN/gMDSCs that are Ly6C+Ly6G+.
Another benefit of viSNE is its reproducibility between analyses. viSNE identified similar immune cell populations in response to either MB49 bladder cancer or B16F10 melanoma. Also, regardless of which cells were used as a starting gate for the viSNE analysis (CD45+, CD45+CD8+ or CD45+Ly6C+Ly6G+), similar populations were consistently generated. These findings indicate that viSNE is useful for evaluating flow cytometry data generated using different tumor models and different pregating strategies.
viSNE distinguished between spleen and tumor Ly6C+Ly6G+ cells based on differential expression of CD11b and CD44. These findings suggested that cells derived from tumors are more activated. This is consistent with reports of CD44 shedding (38) and CD11b upregulation (40) on neutrophils in response to stimulation. However, differences in CD44 expression by granulocytes located in the tumor were ascribed to the effects of enzymes used for tumor dispersal and not necessarily to the inflammatory tumor microenvironment.
viSNE was developed for the analysis of high-dimensional cytometry and has been useful for the analysis of mass cytometry data generated by cytometry by time-of-flight. However, as fluorescence-based flow cytometry has evolved to enable the simultaneous analysis of 18+ colors, high-dimensional analysis tools can also be beneficial for the analysis of traditional flow cytometry. The advantage of high-dimensional analysis is that it is unbiased and takes into consideration all markers to identify cell subsets that may be missed by the use of biaxial gating. Cancer is an example of a disease in which there may be subtle differences in multiple cell populations due to various cues in the tumor microenvironment. Because viSNE uses all parameters to generate an easy-to-visualize three-dimensional plot, the current study suggests that viSNE will prove to be useful for the analysis of tumor immunity.
Supplementary Material
Acknowledgments
We thank Cheryl Kim, director of the La Jolla Institute for Allergy and Immunology flow cytometry core, for instruction on the LSR Fortessa, and for assistance in development of the flow panel with Dr. Caglar Cekic. We thank Amy Blatchley, Dr. Hong Pei, Dr. Yuan Lin, and Dr. Anouk Hamers for assistance with tissue harvesting, and the La Jolla Institute for Allergy and Immunology vivarium technicians for animal care. Additionally, we thank Cheryl Kim, Dr. Erik Ehinger, and Dr. Graham Thomas for helpful discussion on the manuscript.
Abbreviations used in this article
- DC
dendritic cell
- dLN
draining lymph node
- gMDSC
granulocytic myeloid-derived suppressor cell
- MFI
median fluorescent intensity
- MHC II
MHC class II
- PD-1
programmed cell death protein 1
- PMN
polymorphonuclear cell
- SSC
side scatter
- tSNE
t-distributed stochastic neighbor embedding
- viSNE
visualization of t-distributed stochastic neighbor embedding
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Chester C, Maecker HT. Algorithmic tools for mining high-dimensional cytometry data. J Immunol. 2015;195:773–779. doi: 10.4049/jimmunol.1500633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mair F, Hartmann FJ, Mrdjen D, Tosevski V, Krieg C, Becher B. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol. 2016;46:34–43. doi: 10.1002/eji.201545774. [DOI] [PubMed] [Google Scholar]
- 3.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 4.Amir AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 6.Leelatian N, Diggins KE, Irish JM. Characterizing phenotypes and signaling networks of single human cells by mass cytometry. Methods Mol Biol. 2015;1346:99–113. doi: 10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, Irish JM, Kalams SA. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: a comparison of mass and fluorescence cytometry. Cytometry A. 2016;89:271–280. doi: 10.1002/cyto.a.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KWW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 10.Hirakawa M, Matos T, Liu H, Koreth J, Kim HT, Paul NE, Murase K, Whangbo J, Alho AC, Nikiforow S, et al. Low-dose IL-2 selectively activates subsets of CD4+ Tregs and NK cells. JCI Insight. 2016;1:e89278. doi: 10.1172/jci.insight.89278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin L, Frelinger J, Jiang W, Finak G, Seshadri C, Bart PA, Pantaleo G, McElrath J, DeRosa S, Gottardo R. Identification and visualization of multidimensional antigen-specific T-cell populations in polychromatic cytometry data. Cytometry A. 2015;87:675–682. doi: 10.1002/cyto.a.22623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemieux J, Jobin C, Simard C, Néron S. A global look into human T cell subsets before and after cryopreservation using multiparametric flow cytometry and two-dimensional visualization analysis. J Immunol Methods. 2016;434:73–82. doi: 10.1016/j.jim.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Howlader NNA, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, et al. SEER cancer statistics review, 1975–2013. National Cancer Institute; Bethesda, MD: 2015. [Google Scholar]
- 14.White-Gilbertson S, Davis M, Voelkel-Johnson C, Kasman LM. Sex differences in the MB49 syngeneic, murine model of bladder cancer. Bladder. 2016;3:e22. doi: 10.14440/bladder.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Zhang G, Cao Y, Hessner MJ, See WA. MB49 murine urothelial carcinoma: molecular and phenotypic comparison to human cell lines as a model of the direct tumor response to bacillus Calmette-Guerin. J Urol. 2009;182:2932–2937. doi: 10.1016/j.juro.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Günther JH, Jurczok A, Wulf T, Brandau S, Deinert I, Jocham D, Böhle A. Optimizing syngeneic orthotopic murine bladder cancer MB49. Cancer Res. 1999;59:2834–2837. [PubMed] [Google Scholar]
- 17.Loskog A, Ninalga C, Hedlund T, Alimohammadi M, Malmström PU, Tötterman TH. Optimization of the MB49 mouse bladder cancer model for adenoviral gene therapy. Lab Anim. 2005;39:384–393. doi: 10.1258/002367705774286475. [DOI] [PubMed] [Google Scholar]
- 18.Said N, Smith S, Sanchez-Carbayo M, Theodorescu D. Tumor endothelin-1 enhances metastatic colonization of the lung in mouse xenograft models of bladder cancer. J Clin Invest. 2011;121:132–147. doi: 10.1172/JCI42912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Jackson Laboratory. [Accessed: November 1, 2016];Physiological data summary – C57BL/6J (000664) 2007 Available at: http://jackson.jax.org/rs/444-BUH-304/images/physiological_data_000664.pdf.
- 20.Cella M, Engering A, Pinet V, Pieters J, Lanzavecchia A. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature. 1997;388:782–787. doi: 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 21.Itano AA, Jenkins MK. Antigen presentation to naive CD4 T cells in the lymph node. Nat Immunol. 2003;4:733–739. doi: 10.1038/ni957. [DOI] [PubMed] [Google Scholar]
- 22.Wilson NS, El-Sukkari D, Belz GT, Smith CM, Steptoe RJ, Heath WR, Shortman K, Villadangos JA. Most lymphoid organ dendritic cell types are phenotypically and functionally immature. Blood. 2003;102:2187–2194. doi: 10.1182/blood-2003-02-0513. [DOI] [PubMed] [Google Scholar]
- 23.Yao S, Wang S, Zhu Y, Luo L, Zhu G, Flies S, Xu H, Ruff W, Broadwater M, Choi IH, et al. PD-1 on dendritic cells impedes innate immunity against bacterial infection. Blood. 2009;113:5811–5818. doi: 10.1182/blood-2009-02-203141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim TS, Chew V, Sieow JL, Goh S, Yeong JP-S, Soon AL, Ricciardi-Castagnoli P. PD-1 expression on dendritic cells suppresses CD8+ T cell function and antitumor immunity. OncoImmunology. 2015;5:e1085146. doi: 10.1080/2162402X.2015.1085146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114:1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paine A, Kirchner H, Immenschuh S, Oelke M, Blasczyk R, Eiz-Vesper B. IL-2 upregulates CD86 expression on human CD4+ and CD8+ T cells. J Immunol. 2012;188:1620–1629. doi: 10.4049/jimmunol.1100181. [DOI] [PubMed] [Google Scholar]
- 27.Cormier SA, Taranova AG, Bedient C, Nguyen T, Protheroe C, Pero R, Dimina D, Ochkur SI, O’Neill K, Colbert D, et al. Pivotal Advance: eosinophil infiltration of solid tumors is an early and persistent inflammatory host response. J Leukoc Biol. 2006;79:1131–1139. doi: 10.1189/jlb.0106027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carretero R, I, Sektioglu M, Garbi N, Salgado OC, Beckhove P, Hämmerling GJ. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8+ T cells. Nat Immunol. 2015;16:609–617. doi: 10.1038/ni.3159. [DOI] [PubMed] [Google Scholar]
- 29.Davis BP, Rothenberg ME. Eosinophils and cancer. Cancer Immunol Res. 2014;2:1–8. doi: 10.1158/2326-6066.CIR-13-0196. [DOI] [PubMed] [Google Scholar]
- 30.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay J, Tak T, Kamp VM, Koenderman L. Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci. 2013;70:3813–3827. doi: 10.1007/s00018-013-1286-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youn JI, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youn JI, Gabrilovich DI. The biology of myeloid-derived suppressor cells: the blessing and the curse of morphological and functional heterogeneity. Eur J Immunol. 2010;40:2969–2975. doi: 10.1002/eji.201040895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose S, Misharin A, Perlman H. A novel Ly6C/Ly6G-based strategy to analyze the mouse splenic myeloid compartment. Cytometry A. 2012;81:343–350. doi: 10.1002/cyto.a.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sagiv JY, Michaeli J, Assi S, Mishalian I, Kisos H, Levy L, Damti P, Lumbroso D, Polyansky L, Sionov RV, et al. Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Reports. 2015;10:562–573. doi: 10.1016/j.celrep.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 37.Katayama Y, Hidalgo A, Chang J, Peired A, Frenette PS. CD44 is a physiological E-selectin ligand on neutrophils. J Exp Med. 2005;201:1183–1189. doi: 10.1084/jem.20042014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bazil V, Strominger JL. Metalloprotease and serine protease are involved in cleavage of CD43, CD44, and CD16 from stimulated human granulocytes. Induction of cleavage of L-selectin via CD16. J Immunol. 1994;152:1314–1322. [PubMed] [Google Scholar]
- 39.Vremec D, Shortman K. Dendritic cell subtypes in mouse lymphoid organs: cross-correlation of surface markers, changes with incubation, and differences among thymus, spleen, and lymph nodes. J Immunol. 1997;159:565–573. [PubMed] [Google Scholar]
- 40.Wittmann S, Rothe G, Schmitz G, Fröhlich D. Cytokine upregulation of surface antigens correlates to the priming of the neutrophil oxidative burst response. Cytometry A. 2004;57:53–62. doi: 10.1002/cyto.a.10108. [DOI] [PubMed] [Google Scholar]
- 41.Schnorrer P, Behrens GM, Wilson NS, Pooley JL, Smith CM, El-Sukkari D, Davey G, Kupresanin F, Li M, Maraskovsky E, et al. The dominant role of CD8+ dendritic cells in cross-presentation is not dictated by antigen capture. Proc Natl Acad Sci USA. 2006;103:10729–10734. doi: 10.1073/pnas.0601956103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iyoda T, Shimoyama S, Liu K, Omatsu Y, Akiyama Y, Maeda Y, Takahara K, Steinman RM, Inaba K. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J Exp Med. 2002;195:1289–1302. doi: 10.1084/jem.20020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.