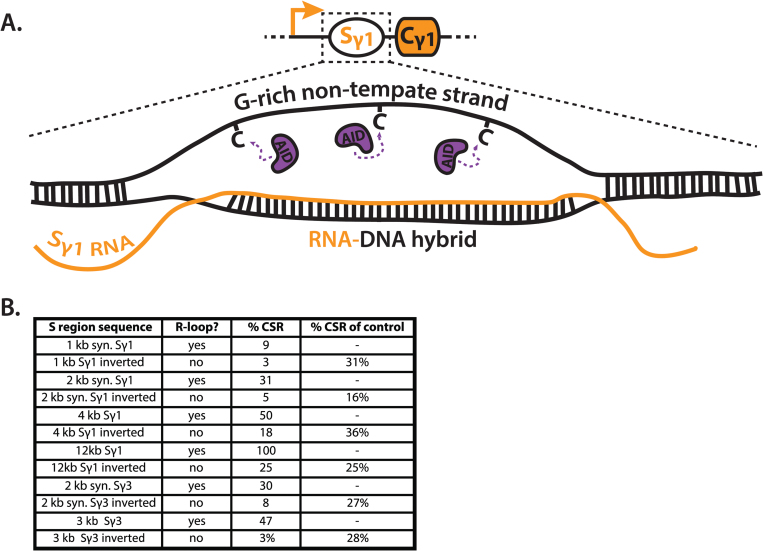
Fig. 4.
R-loops and AID targeting. (A) Transcription through S regions leads to the formation R-loops, consisting of an RNA–DNA hybrid and G-rich non-template single-stranded DNA (ssDNA). R-loops can extend over 1000 bp, as well as several hundred base pairs upstream of S regions. Current models hypothesize the availability of ssDNA is a key factor in targeting activation-induced cytidine deaminase (AID) to R-loops. (B) Table quantifying the contribution of R-loops to class switch recombination. Data were extracted from three studies that compared the ability of various S regions to potentiate IgG1 class switch recombination (CSR) at the endogenous mouse locus, with that of their inverted sequences (74–76). Because the inverted sequences do not support R-loop formation but are identical in size and sequence, an analysis of these comparisons can indicate the contribution of R-loops to CSR. This analysis was posited by Lieber and colleagues (77), and extended to include additional studies. ‘R-loop?’ indicates if the sequence has been shown to form an R-loop. ‘%CSR’ indicates the relative IgG1 CSR frequency, which is defined by the ratio of allele-specific hybridomas produced in various S region replacements, compared with that of the wild-type Sγ1 sequence. ‘%CSR of control’ indicates the frequency of CSR supported by inverted sequences relative to their non-inverted controls. kb, kilobase; syn, synthetic; AID, activation-induced cytidine deaminase.