Abstract
Purpose
This study examines the analytic validity of a software tool designed to provide individuals with risk assessments for colorectal cancer (CRC) based on personal health and family history information. The software is compatible with the U.S. Surgeon General’s My Family Health Portrait.
Methods
An algorithm for risk assessment was created using accepted colorectal risk assessment guidelines, and programmed into a software tool (MFHP). Risk assessments derived from 150 pedigrees using the MFHP tool were compared to “gold standard” risk assessments developed by three expert cancer genetic counselors (GCs).
Results
Genetic counselor risk assessments showed substantial, but not perfect, agreement. MFHP risk assessments for CRC cancer yielded a sensitivity for CRC risk of 81% (95% CI 54–96%) and specificity of 90% (95% CI 83–94%), respectively, when compared to GC pedigree review. The positive predictive values for risk for MFHP was 48% (95% CI 29–68%), while the negative predictive values was 98% (95% CI 93–99%). Agreement between MFHP and GC pedigree review was moderate (Kappa = 0.54)
Conclusions
The analytic validity of the MFHP CRC risk assessment software is similar to other types of screening tools used in primary care. Future investigations should explore the clinical validity and utility of the software in diverse population groups.
Keywords: Family history, colorectal cancer, risk assessment tools, My Family Health Portrait
Introduction
Patient-completed electronic family history (FH) collection tools have been proposed as a way to overcome several of the barriers to collection and use of family history information in health care settings1,2. Several patient completed tools designed to aid in the collection and interpretation of FH have been developed and validated, often in settings such as cancer and primary care clinics3–5. A 2009 review of FH literature conducted for the U.S. Agency for Health Care Research and Quality (AHRQ) and the National Institutes of Health (NIH) identified a paucity of validated FH tools suitable for use in primary care or public health contexts6. In 2014 the National Colorectal Cancer Roundtable found that electronic health record systems (EHRs) are poorly prepared to store and interpret FH relevant to CRC risk assessment7.
The U. S. Centers for Disease Control and Prevention (CDC)-sponsored Family Healthware™ Trial investigated behavioral change related to the clinical use of a patient completed electronic family history tool (FHT) that provided risk assessment for several conditions including, type 2 diabetes, heart disease, stroke, as well as colorectal, breast, and ovarian cancer8. FHT was not designed for stand-alone CRC risk assessment, but extensive expertise was brought to bear in its development, suggesting that FHT may be a useful point of comparison for the evaluation of other automated CRC risk assessment tools. The public version of the My Family Health Portrait (MFHP) tool was developed and launched in 2004 as part of the U.S. Surgeon General’s Family History Initiative. The tool was designed to help the public collect, organize, and share family history information with relatives and health professionals9. The current version of MFHP was designed to collect FH in a format compatible with electronic health record systems using nomenclature and data standards for storing and sharing FH and has been studied in several settings and is freely available to the public10,11,12,13.
An important limitation of the current public MFHP tool is that it does not provide users with feedback or education regarding their individual disease risk. Here we report on the development of a CRC risk assessment module compatible with the public MFHP tool, and, potentially EHRs. As well, we present data regarding the analytic validity of the new module and the FHT tool according to the Analytical validity, Clinical validity; Clinical utility; and Ethical, legal, and social issues (ACCE) framework proposed as a method for evaluating family health history and other genomic health tools14.
Materials and Methods
Creation of the CRC risk module
The MFHP tool output is a structured XML output file that is computable for risk assessment. An algorithm based on National Comprehensive Cancer Network® (NCCN) and United States Preventive Services Task Force (USPSTF) CRC risk assessment guidelines was developed to dichotomize individuals into “elevated” and “not elevated” risk categories based on data entered into the MFHP tool (online appendix A). To avoid inducing potentially inappropriate distress in individuals without benefit of immediate access to evaluation by a health professional, “elevated” risk individuals were not further subdivided into “high” and “moderate” risk categories by the MFHP tool. The model did not incorporate other data elements such as proband gender, body mass index, or self-identified race, as there were no evidence-based guidelines for assigning risk for these factors in conjunction with family history data. The final algorithm was reviewed by a group of cancer risk assessment experts prior to being programmed into an open-source software tool. This input resulted in the final decision to provide a dichotomous rather than a more quantitative risk assessment, and to consider all reported polyps as potential sources of risk warranting further discussion with a health care provider. The output of the MFHP tool is risk specific CRC information printable as letters for public users and their health care providers.
Study population and risk algorithm validation
We used 150 patient-entered pedigrees sequentially derived from the ClinSeq® cohort as the substrate for assessing the MFHP tool and CDC’s FHT CRC module’s analytic performance15. The cohort consists of self-selected volunteers with an interest in learning their personal genomic risks through genome or exome sequencing; individuals entered the study without investigator foreknowledge of cancer family histories. The cohort is more likely to be white, well educated, and affluent than the general U.S. population. Forty additional patient-entered pedigrees derived from the cohort served as internal controls; 20 were deliberately modified to increase colorectal cancer risk to different levels (10 “strong” risk, 10 “moderate” risk) by adding additional accepted risk factors to the pedigree data.
Three different methods were used to derive risk estimates from the test pedigrees: the MFHP tool (including the new CRC risk module, referred to as MFHP hereafter), the CDC’s FHT (1.0 Beta Version for Research Studies), and independent review by three GCs expert in colorectal cancer risk assessment. The GCs were blinded to which pedigrees had been modified to alter risk, the risk assessments made by the MFHP tool, FHT, each other’s risk assessments, and proband self-reported BMI and race. The GCs were instructed that they should categorize the proband’s CRC risk as “weak”, “moderate”, or “strong” according to the criteria outlined by Yoon et al8. This study was approved by the National Human Genome Research Institute Institutional Review Board.
Statistical analysis
Review of pedigrees by a panel of three expert cancer GCs was defined as the gold-standard for risk of CRC in the test pedigrees16. Risk estimates of “weak” by the FHT and GC review were considered “not elevated” while “moderate” and “high” risk assignments were considered to be “elevated”. Pedigrees were scored as showing “elevated” risk for CRC only when two of three counselors reported risk levels as “moderate” or “strong” (true positives). Kappa statistics were generated to assess GC inter-rater agreement for CRC assessment using the “weak”, “moderate” and “strong” risk estimates for all 190 pedigrees. For the calculation of sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV) and Kappa statistics for the presence of CRC risk for the MFHP tool and FHT versus the GC review only the 150 unmodified pedigrees were used.
Results
GC risk estimates for CRC in the ClinSeq® pedigrees
Sixteen of 150 (11%) of the unmodified pedigrees were identified as having “elevated” risk by at least two of three GCs rating risk as “moderate” or “strong”; 6/150 (4%) were identified as having “strong” risk by at least two of three counselors. For the modified internal control pedigrees, the three counselors agreed for 10/10 (100%) and 6/10 (60%) pedigrees intended to be at “strong” or “moderate” risk, respectively. Pair-wise Kappa statistics for the three counselors calculated for all 190 pedigrees were 0.760, 0.776, and 0.851, indicating substantial agreement between the GCs according to the Landis-Koch guidelines17.
Comparison of the MFHP tool versus expert GC risk estimates
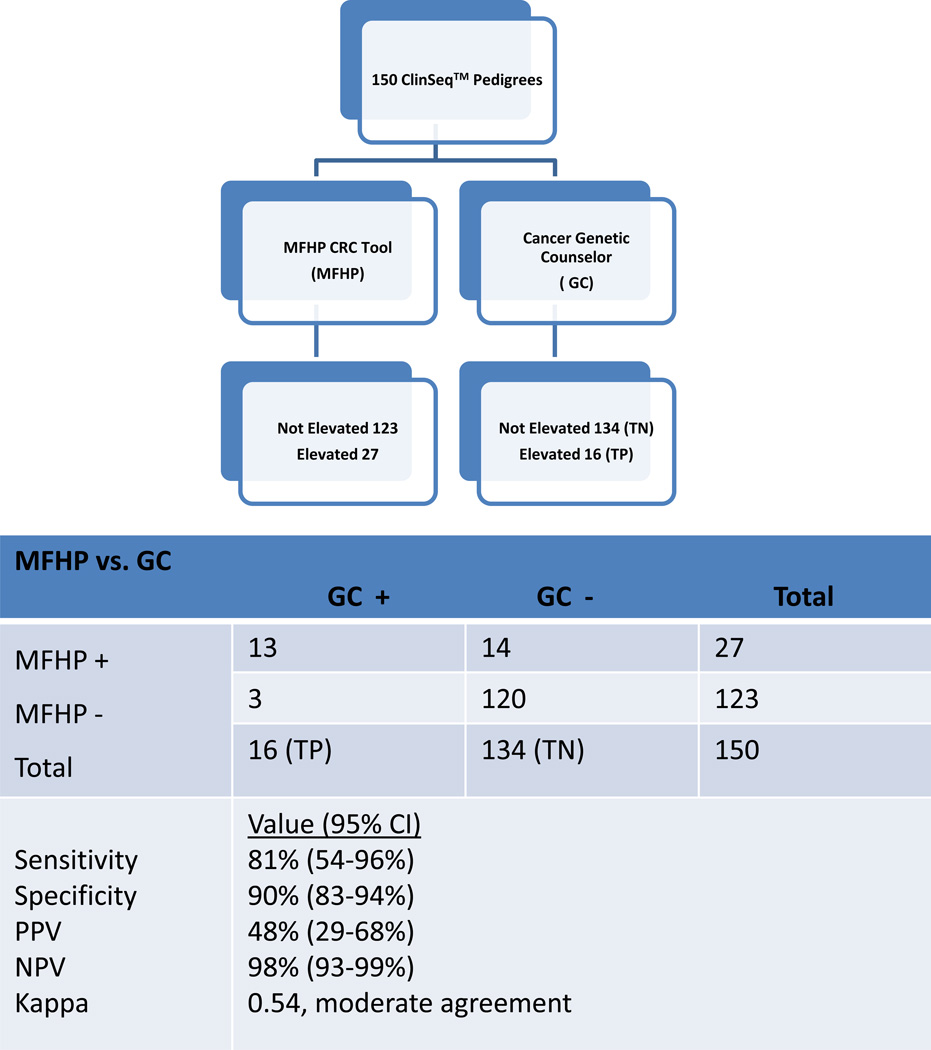
Twenty-seven of 150 (18%) of unmodified pedigrees were assigned “elevated” risk status by the MFHP tool. The MFHP tool had a sensitivity and specificity of 81% (95% CI 54–96%) and 90% (95% CI 83–94%), when compared to pedigrees judged to be “elevated” by GC review. The PPV and NPV were 48% (95% CI 29–68%) and 98%(95% CI 93–99%), respectively (Figure 1). All twenty of the modified pedigrees intended to be at “strong” or “moderate” risk were identified as at “elevated” risk by the MFHP tool. The Kappa statistic between the MFHP tool and GC review suggested moderate agreement (0.54).
Figure 1.
Diagram showing the process used for developing interpretations of 150 unmodified pedigrees derived from the ClinSeq® population (see text) and resulting calculated values for sensitivity, specificity, positive predictive value and negative predictive value calculated Kappa for the My Family Health Portrait colorectal risk algorithm for detection of elevated colorectal cancer risk using expert genetic counselor pedigree interpretation as the “gold standard” for risk interpretation. (TP= true positives; TN= true negatives; PPV= positive predictive value; NPV= negative predictive value; CI= confidence interval)
Comparison of the FHT versus GC risk estimates
Fourteen of 150 (9%) of the unmodified pedigrees were considered to be at “elevated” risk by FHT; of these 14 pedigrees 3/150 (6%) were classified as being at “strong” risk and 11/150 (7%) were rated to be at “moderate” risk. The FHT had a sensitivity and specificity of 75% (95% CI 48–93%)and 99% (95% CI 95–100%) respectively, when compared to pedigrees judged to be “elevated” by GC review. The PPV and NPV for FHT were 86% (95% CI 57–98%) and 97% (95% CI 93–99%) respectively. The Kappa statistic between FHT and GC review indicated substantial agreement (0.78). For the modified control pedigrees there was 10/10 (100%) agreement between the FHT and pedigrees intended to be at “strong” risk. Eight of 10 (80%) of the modified pedigrees intended to be at “moderate” risk were identified as such by FHT, for 2/10 (20%) FHT rated the risk as “strong” instead of “moderate”.
Assessment of disagreement between GC risk estimates and MFHP
There were three unmodified pedigrees that were identified as at “elevated” risk by GC review that were rated “not elevated” risk by the MFHP tool (“false negatives”) and 14 pedigrees for which the MFHP tool rated risk as “elevated” and GC review rated risk as “not elevated” (“false positives”). The sources of the discrepancies were diverse (Table 1). The “false negative” pedigrees shared the characteristic of a lack of CRC cases or colorectal polyps in the proband or relatives. For one of the pedigrees the error source was a failure to enter endometrial cancer as a structured term due to misspelling. The “false positive” pedigrees often appeared to arise from differing assignment of risk arising from personal or family histories of colon polyps or a history of CRC or CRC-associated cancers in second-degree relatives.
Table 1.
Potential sources of heritable risk for colorectal cancer (CRC) present in pedigrees yielding “false negative” and “false positive” results with the My Family Health Portrait colorectal cancer risk assessment (MFHP) tool in comparison to a gold standard of pedigree review by three expert cancer genetic counselors (GC).
| “False negative” | “False positive” |
|---|---|
| GC risk higher than MFHP | MFHP risk higher than GC |
| 2° GasCA; 2° OvCA | 2° CRC; 1° PanCA |
| 1° BrCa, age 40–49; 2° PrCA | 2° CRC, age 40–49 |
| Patient data entry error | 2° with both CRC, BrCA |
| 1° EndoCA, age 30–39 | 2° CRC, age 50–59 |
| 2° CRC; 2° CRC, both age unknown | |
| 2° CRC; 2° CRC, both age unknown | |
| 1° ColP, age 40–49 | |
| 1° GasCA | |
| 2° with both CRC, BrCA | |
| 2° CRC, age 40–49 | |
| 1° AdenoCA; 2° PanCA; 2° PanCA; 2° EndoCA | |
| 1° ColP; 2° ColP; 2° CRC | |
| Proband ColP; 1° ColP | |
| 1° PrCA, 2° GasCA, 2° EndoCA |
Each cell represents a single pedigree, individuals are separated by a semi-colon. Unspecified age ranges for cancer are either unknown or were 60 or greater years of age.
1° – first degree relative; 2° – second degree relative; GasCA – gastric cancer; EndoCA – endometrial/uterine cancer; BrCA – breast cancer; PrCA – prostate cancer; PanCA – pancreatic cancer; ColP – colon polyp; AdenoCA – adenocarcinoma, site unspecified
Discussion
This study provides preliminary evidence supporting the analytic validity of an automated, consumer-oriented screening algorithm for heritable CRC risk assessment that is compatible with the U.S. Surgeon General’s MFHP. To our knowledge it is the first study to formally evaluate the predictive value of a family history risk algorithm designed specifically to work with the MFHP tool. It is important to note that the “gold standard” in this study is not development of disease; rather the risk prediction arrived at by a group of three expert cancer GCs. The PPV values for MFHP and FHT exceed the 10% threshold that is considered an acceptable value for PPV for a genomic screening test18. Because only a fraction of individuals with elevated CRC will actually develop disease, the PPV regarding development of clinical disease is likely to be lower than the values observed in this study, and the NPV higher, for both the MFHP and FHT tools.
The MFHP algorithm was not designed to be diagnostic of CRC syndromes, rather it was designed to be a potential component of an educational tool that helps individuals and their health care providers stratify risk for heritable CRC in order to have an informed conversation regarding further evaluation. As a screening tool intended for use by the general public, it was designed to have a higher sensitivity at the cost of specificity, based on the rationale that missing elevated risk of colon cancer incurs a greater penalty than encouraging additional patients to discuss their risk of colorectal cancer with a health care provider. This choice could be debated, as available evidence suggests family history is not a strong predictor of absolute risk for colorectal cancer in the population19.
Evaluation of the pedigrees for which the risk ratings were discordant between GC review and the MFHP tool suggests that the MFHP tool assigns “elevated” risk to pedigrees with second degree relatives having CRC, multiple Lynch syndrome-related cancers, and personal or family histories of colon polyps. The GCs might not attribute much risk to histories of colon polyps lacking details of pathology. The FHT also does not fully incorporate personal and family history of colon polyps when assigning risk. Assuming such polyps are benign may underestimate risk, and at least one counselor desired additional information20. Two of three instances where the MFHP tool did not assign elevated risk represent somewhat grey areas of risk assessment for CRC. The lack of clarity in guidelines for such grey areas is reflected in the inconsistency in GC risk assignment; for 2 of 3 of the “false negative” pedigrees at least one expert GC rated risk as “weak” while another rated risk as “strong”.
This study has a number of important limitations. Measured internal agreement between the counselors suggests that expert GCs do not always concur when independently assigning CRC risk to pedigrees. The discrepancies between risk estimates provided by GC review and the MFHP algorithm highlight that currently there is no universally accepted “gold standard” for the detection of CRC risk in family histories in routine clinical care. Similar to 2005 study by Qureshi et al., this study uses review by expert GCs to arrive at a proxy gold standard for CRC risk.16 Additional limitations of this study include the small sample size, the nature of the ClinSeq® population, which may not represent the full diversity of the US population, and that the study was only able to examine the analytic validity (and not the clinical utility) of the MFHP CRC algorithm. Future studies should seek to replicate these findings and define the clinical validity and clinical utility of the MFHP tool in additional patient populations.
Supplementary Material
Acknowledgements
The authors thank the following individuals for contributions made to developing the risk assessment algorithm and for their thoughtful comments on the manuscript: Andrew Freedman, Ph.D.; Mitchell Gail, M.D., Ph.D.; Donald Hadley, M.S., C.G.C.; David Lanier, M.D.; Colleen McBride, Ph.D.; Gurvaneet Randhawa, M.D., M.P.H.; Maren Scheuner M.D., M.P.H.; Rodolfo Valdez, Ph.D; and Daniel Wattendorf, M.D., M.S.
Funding: This project was supported by the Genetic Disease Research Branch and the Genomic Healthcare Branch, National Human Genome Research Institute, National Institutes of Health.
Footnotes
Conflict of interest: The authors report no conflict of interest relevant to the content of this manuscript.
Some of the data in this manuscript was presented in abstract form at the 2011 meeting of the American Society of Human Genetics in Montreal, Canada.
References
- 1.Yoon PW, Scheuner MT, Khoury MJ. Research priorities for evaluating family history in the prevention of common chronic diseases. American journal of preventive medicine. 2003 Feb;24(2):128–135. doi: 10.1016/s0749-3797(02)00585-8. [DOI] [PubMed] [Google Scholar]
- 2.Rich EC, Burke W, Heaton CJ, et al. Reconsidering the family history in primary care. Journal of general internal medicine. 2004 Mar;19(3):273–280. doi: 10.1111/j.1525-1497.2004.30401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qureshi N, Carroll JC, Wilson B, et al. The current state of cancer family history collection tools in primary care: a systematic review. Genet Med. 2009 Jul;11(7):495–506. doi: 10.1097/GIM.0b013e3181a7e8e0. [DOI] [PubMed] [Google Scholar]
- 4.Reid GT, Walter FM, Brisbane JM, Emery JD. Family history questionnaires designed for clinical use: a systematic review. Public Health Genomics. 2009;12(2):73–83. doi: 10.1159/000160667. [DOI] [PubMed] [Google Scholar]
- 5.Orlando LA, Buchanan AH, Hahn SE, et al. Development and validation of a primary care-based family health history and decision support program (MeTree) North Carolina medical journal. 2013 Jul-Aug;74(4):287–296. [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi N, Wilson B, Santaguida P, et al. Family history and improving health. Evidence report/technology assessment. 2009 Aug;(186):1–135. [PMC free article] [PubMed] [Google Scholar]
- 7.Special Report: Exome Sequencing for Clinical Diagnosis of Patients with Suspected Genetic Disorders. [Accessed December 19, 2013]; http://www.bcbs.com/blueresources/tec/vols/28/28_03.pdf. [PubMed] [Google Scholar]
- 8.Yoon PW, Scheuner MT, Jorgensen C, Khoury MJ. Developing Family Healthware, a family history screening tool to prevent common chronic diseases. Prev Chronic Dis. 2009 Jan;6(1):A33. [PMC free article] [PubMed] [Google Scholar]
- 9.Facio FM, Feero WG, Linn A, Oden N, Manickam K, Biesecker LG. Validation of My Family Health Portrait for six common heritable conditions. Genet Med. Jun;12(6):370–375. doi: 10.1097/GIM.0b013e3181e15bd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feero WG, Bigley MB, Brinner KM. New standards and enhanced utility for family health history information in the electronic health record: an update from the American Health Information Community's Family Health History Multi-Stakeholder Workgroup. J Am Med Inform Assoc. 2008 Nov-Dec;15(6):723–728. doi: 10.1197/jamia.M2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owens KM, Marvin ML, Gelehrter TD, Ruffin MTt, Uhlmann WR. Clinical use of the Surgeon General's "My Family Health Portrait" (MFHP) tool: opinions of future health care providers. J Genet Couns. Oct;20(5):510–525. doi: 10.1007/s10897-011-9381-x. [DOI] [PubMed] [Google Scholar]
- 12.Kanetzke EE, Lynch J, Prows CA, Siegel RM, Myers MF. Perceived utility of parent-generated family health history as a health promotion tool in pediatric practice. Clin Pediatr (Phila) Aug;50(8):720–728. doi: 10.1177/0009922811403301. [DOI] [PubMed] [Google Scholar]
- 13.The U.S. Surgeon General's My Family Health Portrait. [Accessed July 2, 2012]; https://familyhistory.hhs.gov/fhh-web/home.action.
- 14.Valdez R, Yoon PW, Qureshi N, Green RF, Khoury MJ. Family history in public health practice: a genomic tool for disease prevention and health promotion. Annual review of public health. 2010;31:69–87. doi: 10.1146/annurev.publhealth.012809.103621. 61 p following 87. [DOI] [PubMed] [Google Scholar]
- 15.Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009 Sep;19(9):1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qureshi N, Bethea J, Modell B, et al. Collecting genetic information in primary care: evaluating a new family history tool. Family practice. 2005 Dec;22(6):663–669. doi: 10.1093/fampra/cmi073. [DOI] [PubMed] [Google Scholar]
- 17.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977 Mar;33(1):159–174. [PubMed] [Google Scholar]
- 18.Roberts NJ, Vogelstein JT, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. The predictive capacity of personal genome sequencing. Sci Transl Med. May 9;4(133):133ra158. doi: 10.1126/scitranslmed.3003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor DP, Stoddard GJ, Burt RW, et al. How well does family history predict who will get colorectal cancer? Implications for cancer screening and counseling. Genet Med. May;13(5):385–391. doi: 10.1097/GIM.0b013e3182064384. [DOI] [PubMed] [Google Scholar]
- 20.Madlensky L, Daftary D, Burnett T, et al. Accuracy of colorectal polyp self-reports: findings from the colon cancer family registry. Cancer Epidemiol Biomarkers Prev. 2007 Sep;16(9):1898–1901. doi: 10.1158/1055-9965.EPI-07-0151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.