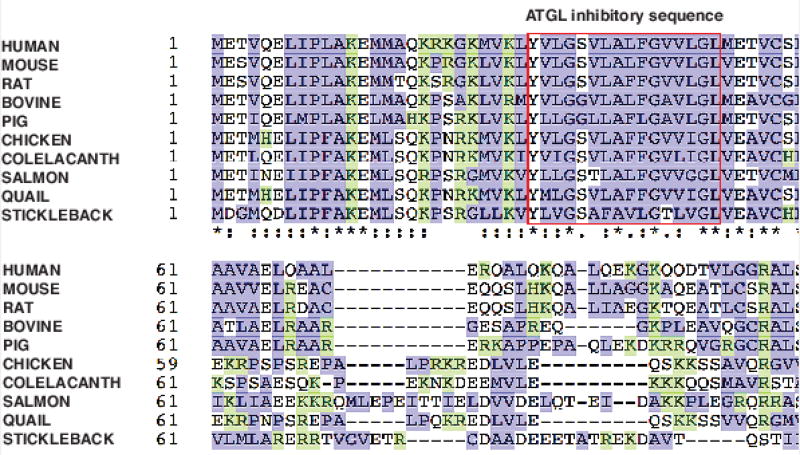
Figure 1. G0S2 protein sequence alignment.
Amino acid sequences of G0S2 proteins from different species were aligned using Clustal Omega program. Overall, the sequences are well conserved with high content of basic (shown in green) and hydrophobic (shown in blue) residues. Red box contains the hydrophobic sequences required for ATGL inhibition. Identical amino acids in all proteins are marked with an asterisk (*), conservative substitutions with a colon (:), and semi-conservative substitutions with a period (.).