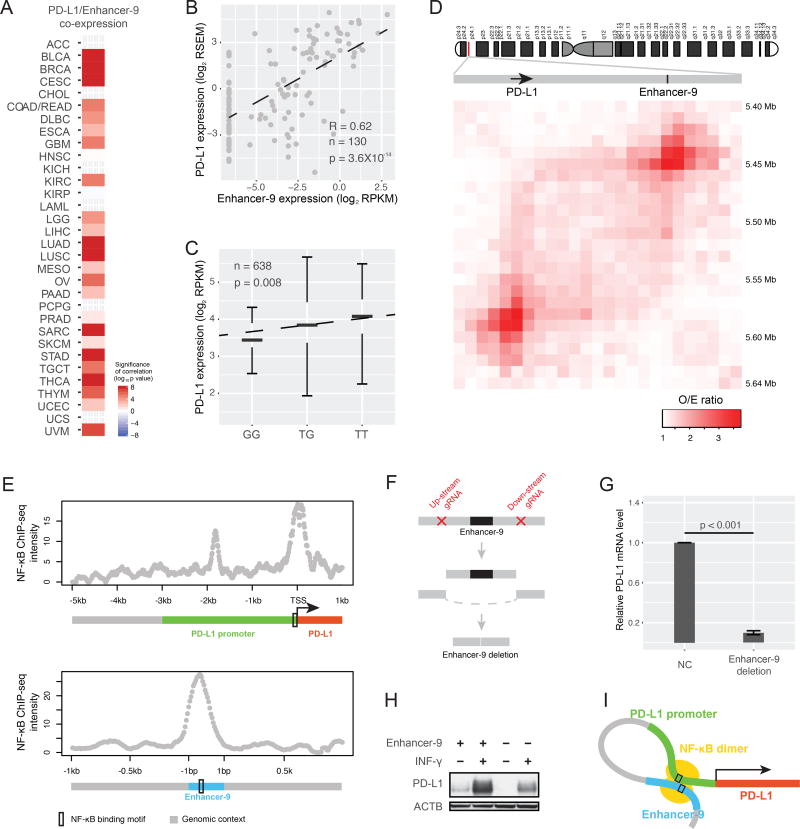
Figure 6. Enhancer-9 regulates PD-L1, a key target of immunotherapy.
(A) Co-expression levels between enhancer-9 (chr9:5580709-5581016) and PD-L1 in multiple cancer types (RNA-seq); p-values for Spearman’s rank correlations were calculated and Bonferroni-corrected. (B) Scatter plot showing co-expression between PD-L1 mRNA level and enhancer-9 expression level. (C) SNP rs1536927 near enhancer 9 is a PD-L1 eQTL; p-value was calculated using ANOVA. (D) Direct interaction between PD-L1 gene body and enhancer 9 detected by Hi-C. The Hi-C O/E ratio was calculated as the median of O/E ratios of 7 human cell lines. (E) NF-κB ChIP-seq signals of enhancer-9 and the PD-L1 promoter. (F) Experimental design of sgRNA-guided enhancer perturbation by Cas9 protein. Three different sgRNAs were designed for each side of the enhancer. (G) Relative mRNA expression levels of PD-L1 in A549 cells and the same line after homozygous enhancer-9 deletion. Error bars show mean ± SE of results of 4 replicates; the difference was assessed using t-test. (H) PD-L1 protein levels in the control and enhancer-9 deletion cell lines without and with INF-γ stimulation. (I) Cartoon of NF-κB-mediated enhancer/promoter interaction for PD-L1 activation. See also Figure S6.