Figure 2. Overview of ArM evolution protocol.
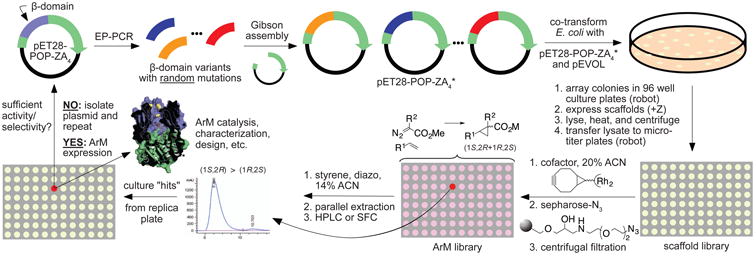
From Top-Left: The gene encoding a POP scaffold (e.g. 0-ZA4) in pET-28 (the POP β-domain is shown in grey-blue in both plasmid and the ArM structure model) is used to generate a library of β-domain variants with random mutations via error prone PCR. The remaining backbone is amplified separately, and the gene library is ligated via Gibson assembly. The gene library is co-transformed into E. coli with pEVOL-pAzF, a plasmid containing an orthogonal tRNA and aaRS for Amber stop codon suppression. A colony picker robot is used to array colonies into 96 well plates, where the POP genes are expressed, and cells are lysed, heated, and centrifuged. Next, a liquid handling robot is used to transfer the lysate to a fresh 96 well plate, cofactor is added to generate POP ArMs, and an azide-substituted resin is added to scavenge unreacted cofactor. The ARM library is screened by HPLC or SFC for increased enantioselectivity of olefin cyclopropanation. Putative hits are isolated and cultured for verification on a larger scale without the presence of cellular lysate debris. If the putative hit is validated, it is used as parent for an additional round of mutagenesis until the desired activity/selectivity is observed.
