Figure 5. MYO6 is linked to the RhoGEF DOCK7 via LRCH3.
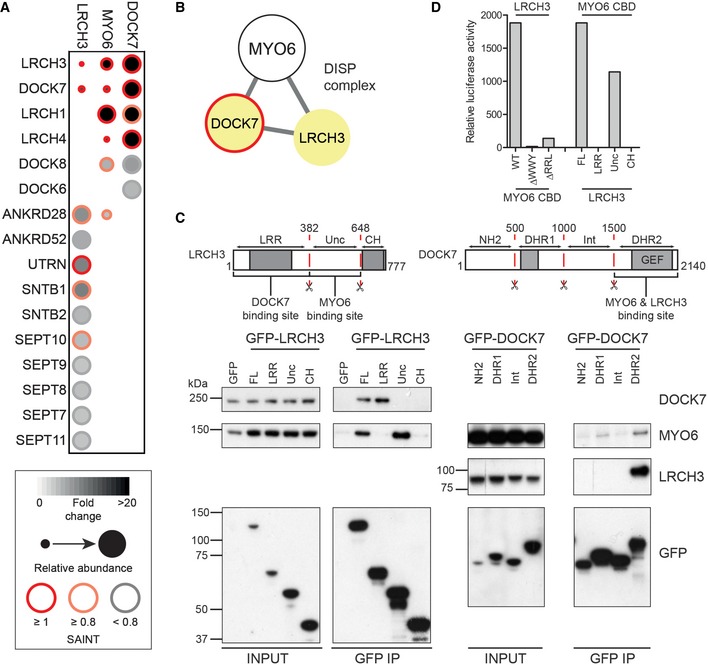
- Dot plot of high and medium confidence interactions (> 3 FC‐A and > 0.8 SAINT or > 3 FC‐A and < 0.8 SAINT) identified in BirA*‐LRCH3 and BirA*‐DOCK7 experiments and shared interactors from the BirA*‐MYO6 CBD interactome.
- Network diagram of the DISP complex.
- Top: Schematic cartoon highlighting domain structure, fragments and binding sites found in LRCH3 (left) and DOCK7 (right). Bottom: GFP nanobody immunoprecipitates from HEK293T cells transfected with GFP, full‐length GFP‐LRCH3 and GFP‐LRCH3 fragments corresponding to amino acids 1–382 (LRR), 383–648 (Unc) or 649–777 (CH) or GFP‐DOCK7 fragments corresponding to amino acids 1–500 (NH2), 501–1000 (DHR1), 1001–1500 (Int) or 1501–2140 (DHR2). Samples were analysed by Western blot with the indicated antibodies.
- The mammalian two‐hybrid assay was used to test direct binding of full‐length LRCH3 and wild‐type, ΔWWY or ΔRRL MYO6 tail and full‐length LRCH3 or LRCH3 fragments and wild‐type MYO6 tail. Graph shows relative luciferase activity from a single representative experiment.
Source data are available online for this figure.
