Figure 2.
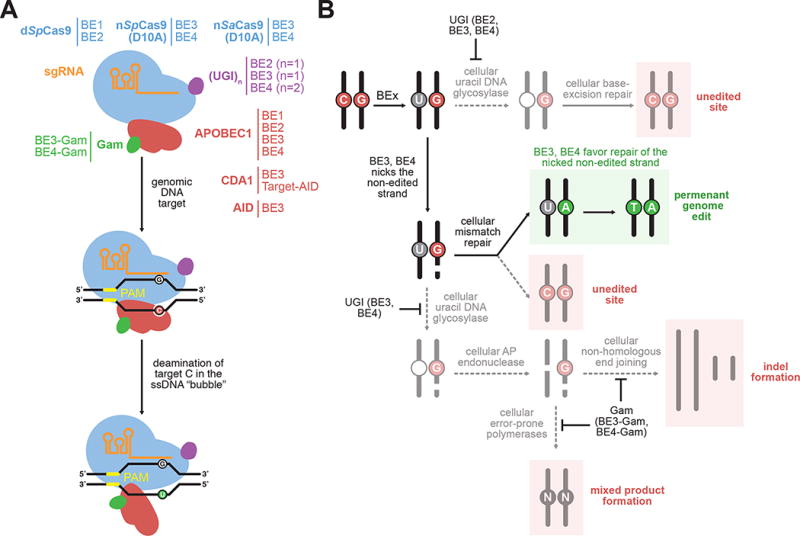
Overview of C•G-to-T•A base editing. (A) The components of C•G-to-T•A base editors. The sgRNA (orange) complexes with a Cas9 homolog (blue) to direct the base editor to a genomic locus of interest. A ssDNA-specific cytidine deaminase enzyme (red) catalytically deaminates cytidine nucleobases within the single-stranded portion of the R-loop to uracil nucleobases. The addition of one or more copies of uracil glycosylase inhibitor (UGI, purple) and the bacteriophage Mu DSB-binding protein Gam (green) help to maximize resolution of the U•G intermediate to a T•A base pair. (B) Possible outcomes following formation of the U•G base editing intermediate. The base excision repair protein uracil DNA glycosylase (UNG) can excise uracil from the U•G mismatch, resulting in an abasic site that is ultimately converted back to a C•G base pair. The UGI component of base editors (in BE2, BE3, and BE4) suppresses uracil excision. The BE3 and BE4 editors incorporate a Cas9 nickase to selectively nick the G-containing strand opposite the converted uracil. This nick guides the mismatch repair machinery to preferentially replace the G-containing strand, resulting in the desired T•A base pair. Excision of uracil from the nick-containing intermediate by UNG, followed by abasic site processing by AP endonuclease can yield a DSB. The DSB can be processed by NHEJ to produce indels, or by error-prone polymerases to yield mixed products such as G•C or A•T base pairs.
