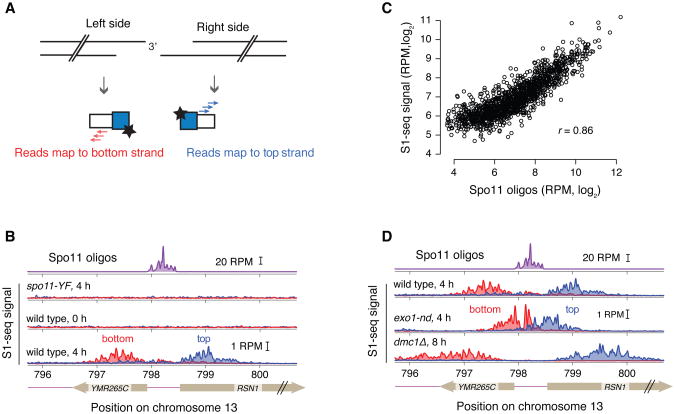
Figure 3.
Validation of S1-seq: A. Schematic of the S1-seq signal polarity. Resection to the right or left should yield reads mapping to the top or bottom strands, respectively. B. An example of Spo11-oligo and strand-specific S1-seq reads (51-bp smoothing) surrounding the RSN1 hotspot. C. S1-seq correlates well with Spo11 oligos: S1-seq signal (wild-type, 4 h) was summed from +200 to +1600 (top strand) and -200 to -1600 (bottom strand) relative to hotspot midpoints and plotted against the sum of the Spo11-oligo hitcount for each corresponding hotspot. For this analysis, hotspots with no other hotspot within 2 kb were used. D. Spo11-oligo and strand-specific S1-seq reads as in panel B. Panels B, C, D reproduced from Mimitou, Yamada, & Keeney, 2017.